Efficient dual-negative selection for bacterial genome editing
- PMID: 32448155
- PMCID: PMC7245781
- DOI: 10.1186/s12866-020-01819-2
Efficient dual-negative selection for bacterial genome editing
Abstract
Background: Gene editing is key for elucidating gene function. Traditional methods, such as consecutive single-crossovers, have been widely used to modify bacterial genomes. However, cumbersome cloning and limited efficiency of negative selection often make this method slower than other methods such as recombineering.
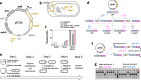
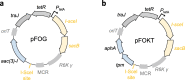
Results: Here, we established a time-effective variant of consecutive single-crossovers. This method exploits rapid plasmid construction using Gibson assembly, a convenient E. coli donor strain, and efficient dual-negative selection for improved suicide vector resolution. We used this method to generate in-frame deletions, insertions and point mutations in Salmonella enterica with limited hands-on time. Adapted versions enabled efficient gene editing also in Pseudomonas aeruginosa and multi-drug resistant (MDR) Escherichia coli clinical isolates.
Conclusions: Our method is time-effective and allows facile manipulation of multiple bacterial species including MDR clinical isolates. We anticipate that this method might be broadly applicable to additional bacterial species, including those for which recombineering has been difficult to implement.
Keywords: Gene manipulation; Homologous recombination; MDR; Mutagenesis; Salmonella.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
A versatile and highly efficient method for scarless genome editing in Escherichia coli and Salmonella enterica.BMC Biotechnol. 2014 Sep 25;14:84. doi: 10.1186/1472-6750-14-84. BMC Biotechnol. 2014. PMID: 25255806 Free PMC article.
-
A rapid seamless method for gene knockout in Pseudomonas aeruginosa.BMC Microbiol. 2017 Sep 19;17(1):199. doi: 10.1186/s12866-017-1112-5. BMC Microbiol. 2017. PMID: 28927382 Free PMC article.
-
Combination of ssDNA recombineering and CRISPR-Cas9 for Pseudomonas putida KT2440 genome editing.Appl Microbiol Biotechnol. 2019 Mar;103(6):2783-2795. doi: 10.1007/s00253-019-09654-w. Epub 2019 Feb 14. Appl Microbiol Biotechnol. 2019. PMID: 30762073
-
An expanded CRISPR-Cas9-assisted recombineering toolkit for engineering genetically intractable Pseudomonas aeruginosa isolates.Nat Protoc. 2023 Nov;18(11):3253-3288. doi: 10.1038/s41596-023-00882-z. Epub 2023 Oct 5. Nat Protoc. 2023. PMID: 37798358 Review.
-
CRISPR/Cas9-Based Counterselection Boosts Recombineering Efficiency in Pseudomonas putida.Biotechnol J. 2018 May;13(5):e1700161. doi: 10.1002/biot.201700161. Epub 2017 Dec 4. Biotechnol J. 2018. PMID: 29058367 Review.
Cited by
-
Characterization of Pseudomonas aeruginosa resistance to ceftolozane-tazobactam due to ampC and/or ampD mutations observed during treatment using semi-mechanistic PKPD modeling.Antimicrob Agents Chemother. 2023 Oct 18;67(10):e0048023. doi: 10.1128/aac.00480-23. Epub 2023 Sep 11. Antimicrob Agents Chemother. 2023. PMID: 37695298 Free PMC article.
-
CRISPR/Cas9-Mediated Genome Editing for Pseudomonas fulva, a Novel Pseudomonas Species with Clinical, Animal, and Plant-Associated Isolates.Int J Mol Sci. 2022 May 13;23(10):5443. doi: 10.3390/ijms23105443. Int J Mol Sci. 2022. PMID: 35628253 Free PMC article.
-
Transforming the untransformable with knockout minicircles: High-efficiency transformation and vector-free allelic exchange knockout in the fish pathogen Photobacterium damselae.Microbiologyopen. 2023 Aug;12(4):e1374. doi: 10.1002/mbo3.1374. Microbiologyopen. 2023. PMID: 37642481 Free PMC article.
-
High-throughput fitness experiments reveal specific vulnerabilities of human-adapted Salmonella during stress and infection.Nat Genet. 2024 Jun;56(6):1288-1299. doi: 10.1038/s41588-024-01779-7. Epub 2024 Jun 3. Nat Genet. 2024. PMID: 38831009 Free PMC article.
-
Tissue compartmentalization enables Salmonella persistence during chemotherapy.Proc Natl Acad Sci U S A. 2021 Dec 21;118(51):e2113951118. doi: 10.1073/pnas.2113951118. Proc Natl Acad Sci U S A. 2021. PMID: 34911764 Free PMC article.
References
-
- Camps M, Loeb LA. Targeted mutagenesis in E. coli: a powerful tool for the generation of random mutant libraries. Discov Med. 2003;3(18):36–37. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

