TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes
- PMID: 32404436
- PMCID: PMC7285829
- DOI: 10.1126/sciimmunol.abc3582
TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes
Abstract
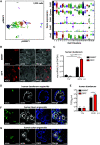
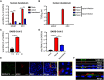
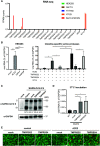
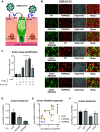
Gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA are frequently observed in COVID-19 patients. However, it is unclear whether SARS-CoV-2 replicates in the human intestine and contributes to possible fecal-oral transmission. Here, we report productive infection of SARS-CoV-2 in ACE2+ mature enterocytes in human small intestinal enteroids. Expression of two mucosa-specific serine proteases, TMPRSS2 and TMPRSS4, facilitated SARS-CoV-2 spike fusogenic activity and promoted virus entry into host cells. We also demonstrate that viruses released into the intestinal lumen were inactivated by simulated human colonic fluid, and infectious virus was not recovered from the stool specimens of COVID-19 patients. Our results highlight the intestine as a potential site of SARS-CoV-2 replication, which may contribute to local and systemic illness and overall disease progression.
Copyright © 2020, The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures

Similar articles
-
Relative Abundance of SARS-CoV-2 Entry Genes in the Enterocytes of the Lower Gastrointestinal Tract.Genes (Basel). 2020 Jun 11;11(6):645. doi: 10.3390/genes11060645. Genes (Basel). 2020. PMID: 32545271 Free PMC article.
-
SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor.Cell. 2020 Apr 16;181(2):271-280.e8. doi: 10.1016/j.cell.2020.02.052. Epub 2020 Mar 5. Cell. 2020. PMID: 32142651 Free PMC article.
-
Expression profiling meta-analysis of ACE2 and TMPRSS2, the putative anti-inflammatory receptor and priming protease of SARS-CoV-2 in human cells, and identification of putative modulators.Redox Biol. 2020 Sep;36:101615. doi: 10.1016/j.redox.2020.101615. Epub 2020 Jun 24. Redox Biol. 2020. PMID: 32863223 Free PMC article.
-
Targeting the intestinal TMPRSS2 protease to prevent SARS-CoV-2 entry into enterocytes-prospects and challenges.Mol Biol Rep. 2021 May;48(5):4667-4675. doi: 10.1007/s11033-021-06390-1. Epub 2021 May 22. Mol Biol Rep. 2021. PMID: 34023987 Free PMC article. Review.
-
COVID-19, coronavirus, SARS-CoV-2 and the small bowel.Rev Esp Enferm Dig. 2020 May;112(5):383-388. doi: 10.17235/reed.2020.7137/2020. Rev Esp Enferm Dig. 2020. PMID: 32343593 Review.
Cited by
-
Bivalent SARS-CoV-2 mRNA vaccines increase breadth of neutralization and protect against the BA.5 Omicron variant.bioRxiv [Preprint]. 2022 Sep 13:2022.09.12.507614. doi: 10.1101/2022.09.12.507614. bioRxiv. 2022. Update in: Nat Med. 2023 Jan;29(1):247-257. doi: 10.1038/s41591-022-02092-8 PMID: 36263060 Free PMC article. Updated. Preprint.
-
COVID-19 infection in severe Alpha 1-antitrypsin deficiency: Looking for a rationale.Respir Med. 2021 Jul;183:106440. doi: 10.1016/j.rmed.2021.106440. Epub 2021 Apr 30. Respir Med. 2021. PMID: 33964815 Free PMC article. Review.
-
Deciphering COVID-19 host transcriptomic complexity and variations for therapeutic discovery against new variants.iScience. 2022 Oct 21;25(10):105068. doi: 10.1016/j.isci.2022.105068. Epub 2022 Sep 3. iScience. 2022. PMID: 36093376 Free PMC article.
-
Immunogenicity and efficacy of XBB.1.5 rS vaccine against the EG.5.1 variant of SARS-CoV-2 in Syrian hamsters.J Virol. 2024 Oct 22;98(10):e0052824. doi: 10.1128/jvi.00528-24. Epub 2024 Sep 4. J Virol. 2024. PMID: 39230305
-
Severe pediatric COVID-19: a review from the clinical and immunopathophysiological perspectives.World J Pediatr. 2024 Apr;20(4):307-324. doi: 10.1007/s12519-023-00790-y. Epub 2024 Feb 6. World J Pediatr. 2024. PMID: 38321331 Free PMC article. Review.
References
-
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., Zhang Y.-Z., A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). 10.1038/s41586-020-2008-3 - DOI - PMC - PubMed
-
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). 10.1038/s41586-020-2012-7 - DOI - PMC - PubMed
-
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.-H., Nitsche A., Müller M. A., Drosten C., Pöhlmann S., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280.e8 (2020). 10.1016/j.cell.2020.02.052 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 75N93019C00062/AI/NIAID NIH HHS/United States
- R00 AI135031/AI/NIAID NIH HHS/United States
- K99 AI135031/AI/NIAID NIH HHS/United States
- F32 AI138392/AI/NIAID NIH HHS/United States
- TL1 TR002344/TR/NCATS NIH HHS/United States
- S10 OD020141/OD/NIH HHS/United States
- R01 AI150796/AI/NIAID NIH HHS/United States
- R37 AI059371/AI/NIAID NIH HHS/United States
- KL2 TR002346/TR/NCATS NIH HHS/United States
- R01 DK109384/DK/NIDDK NIH HHS/United States
- UL1 TR002345/TR/NCATS NIH HHS/United States
- R01 AI127828/AI/NIAID NIH HHS/United States
- R01 AI125249/AI/NIAID NIH HHS/United States
- R01 AI059371/AI/NIAID NIH HHS/United States
- P30 DK052574/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

