Transcriptional and Cellular Diversity of the Human Heart
- PMID: 32403949
- PMCID: PMC7666104
- DOI: 10.1161/CIRCULATIONAHA.119.045401
Transcriptional and Cellular Diversity of the Human Heart
Abstract
Background: The human heart requires a complex ensemble of specialized cell types to perform its essential function. A greater knowledge of the intricate cellular milieu of the heart is critical to increase our understanding of cardiac homeostasis and pathology. As recent advances in low-input RNA sequencing have allowed definitions of cellular transcriptomes at single-cell resolution at scale, we have applied these approaches to assess the cellular and transcriptional diversity of the nonfailing human heart.
Methods: Microfluidic encapsulation and barcoding was used to perform single nuclear RNA sequencing with samples from 7 human donors, selected for their absence of overt cardiac disease. Individual nuclear transcriptomes were then clustered based on transcriptional profiles of highly variable genes. These clusters were used as the basis for between-chamber and between-sex differential gene expression analyses and intersection with genetic and pharmacologic data.
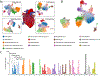
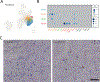
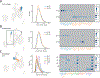
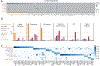
Results: We sequenced the transcriptomes of 287 269 single cardiac nuclei, revealing 9 major cell types and 20 subclusters of cell types within the human heart. Cellular subclasses include 2 distinct groups of resident macrophages, 4 endothelial subtypes, and 2 fibroblast subsets. Comparisons of cellular transcriptomes by cardiac chamber or sex reveal diversity not only in cardiomyocyte transcriptional programs but also in subtypes involved in extracellular matrix remodeling and vascularization. Using genetic association data, we identified strong enrichment for the role of cell subtypes in cardiac traits and diseases. Intersection of our data set with genes on cardiac clinical testing panels and the druggable genome reveals striking patterns of cellular specificity.
Conclusions: Using large-scale single nuclei RNA sequencing, we defined the transcriptional and cellular diversity in the normal human heart. Our identification of discrete cell subtypes and differentially expressed genes within the heart will ultimately facilitate the development of new therapeutics for cardiovascular diseases.
Keywords: RNA; cardiovascular disease; heart.
Figures


Similar articles
-
Cells of the adult human heart.Nature. 2020 Dec;588(7838):466-472. doi: 10.1038/s41586-020-2797-4. Epub 2020 Sep 24. Nature. 2020. PMID: 32971526 Free PMC article.
-
Single-Cell Reconstruction of Progression Trajectory Reveals Intervention Principles in Pathological Cardiac Hypertrophy.Circulation. 2020 May 26;141(21):1704-1719. doi: 10.1161/CIRCULATIONAHA.119.043053. Epub 2020 Feb 26. Circulation. 2020. PMID: 32098504
-
Single-Cell Resolution of Temporal Gene Expression during Heart Development.Dev Cell. 2016 Nov 21;39(4):480-490. doi: 10.1016/j.devcel.2016.10.001. Epub 2016 Nov 10. Dev Cell. 2016. PMID: 27840107 Free PMC article.
-
Dynamic cell-cell and cell-ECM interactions in the heart.J Mol Cell Cardiol. 2014 May;70:19-26. doi: 10.1016/j.yjmcc.2013.10.006. Epub 2013 Oct 17. J Mol Cell Cardiol. 2014. PMID: 24140801 Review.
-
Origin of cardiac fibroblasts and the role of periostin.Circ Res. 2009 Nov 6;105(10):934-47. doi: 10.1161/CIRCRESAHA.109.201400. Circ Res. 2009. PMID: 19893021 Free PMC article. Review.
Cited by
-
PLXND1-mediated calcium dyshomeostasis impairs endocardial endothelial autophagy in atrial fibrillation.Front Physiol. 2022 Aug 9;13:960480. doi: 10.3389/fphys.2022.960480. eCollection 2022. Front Physiol. 2022. PMID: 36017337 Free PMC article.
-
Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell-cell interactions.Cell Rep Med. 2021 May 18;2(5):100287. doi: 10.1016/j.xcrm.2021.100287. Epub 2021 May 3. Cell Rep Med. 2021. PMID: 33969320 Free PMC article.
-
Inflammatory signalling in atrial cardiomyocytes: a novel unifying principle in atrial fibrillation pathophysiology.Nat Rev Cardiol. 2023 Mar;20(3):145-167. doi: 10.1038/s41569-022-00759-w. Epub 2022 Sep 15. Nat Rev Cardiol. 2023. PMID: 36109633 Free PMC article. Review.
-
Sex-specific responses to slow progressive pressure overload in a large animal model of HFpEF.Am J Physiol Heart Circ Physiol. 2022 Oct 1;323(4):H797-H817. doi: 10.1152/ajpheart.00374.2022. Epub 2022 Sep 2. Am J Physiol Heart Circ Physiol. 2022. PMID: 36053749 Free PMC article.
-
Single Nucleus RNA-sequencing Reveals Altered Intercellular Communication and Dendritic Cell Activation in Nonobstructive Hypertrophic Cardiomyopathy.Cardiol Cardiovasc Med. 2022;6(4):398-415. doi: 10.26502/fccm.92920277. Epub 2022 Aug 19. Cardiol Cardiovasc Med. 2022. PMID: 36237479 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

