A serological assay to detect SARS-CoV-2 seroconversion in humans
- PMID: 32398876
- PMCID: PMC8183627
- DOI: 10.1038/s41591-020-0913-5
A serological assay to detect SARS-CoV-2 seroconversion in humans
Abstract
Here, we describe a serological enzyme-linked immunosorbent assay for the screening and identification of human SARS-CoV-2 seroconverters. This assay does not require the handling of infectious virus, can be adjusted to detect different antibody types in serum and plasma and is amenable to scaling. Serological assays are of critical importance to help define previous exposure to SARS-CoV-2 in populations, identify highly reactive human donors for convalescent plasma therapy and investigate correlates of protection.
Conflict of interest statement
Competing interests
Mount Sinai is in the process of licensing out assays to commercial entities based on the assays described here and has filed for patent protection.
Figures


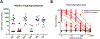
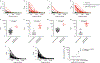

Update of
-
A serological assay to detect SARS-CoV-2 seroconversion in humans.medRxiv [Preprint]. 2020 Apr 16:2020.03.17.20037713. doi: 10.1101/2020.03.17.20037713. medRxiv. 2020. Update in: Nat Med. 2020 Jul;26(7):1033-1036. doi: 10.1038/s41591-020-0913-5. PMID: 32511441 Free PMC article. Updated. Preprint.
Comment in
-
The serostatus approach to fighting COVID-19.Int J Infect Dis. 2020 May;94:53-54. doi: 10.1016/j.ijid.2020.03.080. Epub 2020 Apr 7. Int J Infect Dis. 2020. PMID: 32276044 Free PMC article. No abstract available.
Similar articles
-
A comparison of four serological assays for detecting anti-SARS-CoV-2 antibodies in human serum samples from different populations.Sci Transl Med. 2020 Sep 2;12(559):eabc3103. doi: 10.1126/scitranslmed.abc3103. Epub 2020 Aug 17. Sci Transl Med. 2020. PMID: 32817357 Free PMC article.
-
Analysis of COVID-19 convalescent plasma for SARS-CoV-2 IgG using two commercial immunoassays.J Immunol Methods. 2020 Nov;486:112837. doi: 10.1016/j.jim.2020.112837. Epub 2020 Aug 20. J Immunol Methods. 2020. PMID: 32828791 Free PMC article.
-
Convalescent plasma for COVID-19: Promising, not proven.Cleve Clin J Med. 2020 Nov 2;87(11):664-670. doi: 10.3949/ccjm.87a.ccc056. Cleve Clin J Med. 2020. PMID: 32759176
-
Convalescent plasma therapy for COVID-19: a tried-and-true old strategy?Signal Transduct Target Ther. 2020 Sep 15;5(1):203. doi: 10.1038/s41392-020-00310-8. Signal Transduct Target Ther. 2020. PMID: 32934211 Free PMC article. Review. No abstract available.
-
COVID-19: an Immunopathological View.mSphere. 2020 Apr 22;5(2):e00344-20. doi: 10.1128/mSphere.00344-20. mSphere. 2020. PMID: 32321823 Free PMC article. Review.
Cited by
-
Engineered protein subunit COVID19 vaccine is as immunogenic as nanoparticles in mouse and hamster models.Sci Rep. 2024 Oct 26;14(1):25528. doi: 10.1038/s41598-024-76377-y. Sci Rep. 2024. PMID: 39462119 Free PMC article.
-
Implementation of an Immunoassay Based on the MVA-T7pol-Expression System for Rapid Identification of Immunogenic SARS-CoV-2 Antigens: A Proof-of-Concept Study.Int J Mol Sci. 2024 Oct 10;25(20):10898. doi: 10.3390/ijms252010898. Int J Mol Sci. 2024. PMID: 39456680 Free PMC article.
-
Three doses of Sars-CoV-2 mRNA vaccine in older adults result in similar antibody responses but reduced cellular cytokine responses relative to younger adults.Vaccine X. 2024 Sep 25;20:100564. doi: 10.1016/j.jvacx.2024.100564. eCollection 2024 Oct. Vaccine X. 2024. PMID: 39403561 Free PMC article.
-
Optimization of Cellular Transduction by the HIV-Based Pseudovirus Platform with Pan-Coronavirus Spike Proteins.Viruses. 2024 Sep 20;16(9):1492. doi: 10.3390/v16091492. Viruses. 2024. PMID: 39339968 Free PMC article.
-
Evaluating Immunologic and Illness Outcomes of SARS-CoV-2 Infection in Vaccinated and Unvaccinated Children Aged ≥ 5 Years, in a Multisite Longitudinal Cohort.Diseases. 2024 Aug 1;12(8):171. doi: 10.3390/diseases12080171. Diseases. 2024. PMID: 39195170 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

