The Lysosomotropic Activity of Hydrophobic Weak Base Drugs is Mediated via Their Intercalation into the Lysosomal Membrane
- PMID: 32349204
- PMCID: PMC7290590
- DOI: 10.3390/cells9051082
The Lysosomotropic Activity of Hydrophobic Weak Base Drugs is Mediated via Their Intercalation into the Lysosomal Membrane
Abstract
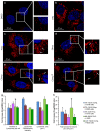
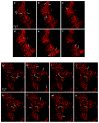
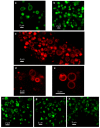
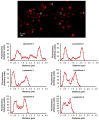
Lipophilic weak base therapeutic agents, termed lysosomotropic drugs (LDs), undergo marked sequestration and concentration within lysosomes, hence altering lysosomal functions. This lysosomal drug entrapment has been described as luminal drug compartmentalization. Consistent with our recent finding that LDs inflict a pH-dependent membrane fluidization, we herein demonstrate that LDs undergo intercalation and concentration within lysosomal membranes. The latter was revealed experimentally and computationally by (a) confocal microscopy of fluorescent compounds and drugs within lysosomal membranes, and (b) molecular dynamics modeling of the pH-dependent membrane insertion and accumulation of an assortment of LDs, including anticancer drugs. Based on the multiple functions of the lysosome as a central nutrient sensory hub and a degradation center, we discuss the molecular mechanisms underlying the alteration of morphology and impairment of lysosomal functions as consequences of LDs' intercalation into lysosomes. Our findings bear important implications for drug design, drug induced lysosomal damage, diseases and pertaining therapeutics.
Keywords: drug sequestration; lysosomes; lysosomotropic drugs; membrane intercalation; molecular dynamics.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
Similar articles
-
Lysosomal accumulation of anticancer drugs triggers lysosomal exocytosis.Oncotarget. 2017 Jul 11;8(28):45117-45132. doi: 10.18632/oncotarget.15155. Oncotarget. 2017. PMID: 28187461 Free PMC article.
-
Lysosomal sequestration of hydrophobic weak base chemotherapeutics triggers lysosomal biogenesis and lysosome-dependent cancer multidrug resistance.Oncotarget. 2015 Jan 20;6(2):1143-56. doi: 10.18632/oncotarget.2732. Oncotarget. 2015. PMID: 25544758 Free PMC article.
-
Drug Sequestration in Lysosomes as One of the Mechanisms of Chemoresistance of Cancer Cells and the Possibilities of Its Inhibition.Int J Mol Sci. 2020 Jun 20;21(12):4392. doi: 10.3390/ijms21124392. Int J Mol Sci. 2020. PMID: 32575682 Free PMC article. Review.
-
Lysosomes as mediators of drug resistance in cancer.Drug Resist Updat. 2016 Jan;24:23-33. doi: 10.1016/j.drup.2015.11.004. Epub 2015 Nov 26. Drug Resist Updat. 2016. PMID: 26830313 Review.
-
Lysosomotropic drugs activate TFEB via lysosomal membrane fluidization and consequent inhibition of mTORC1 activity.Cell Death Dis. 2018 Dec 13;9(12):1191. doi: 10.1038/s41419-018-1227-0. Cell Death Dis. 2018. PMID: 30546014 Free PMC article.
Cited by
-
Recent advances on molecular dynamics-based techniques to address drug membrane permeability with atomistic detail.BBA Adv. 2023 Aug 16;4:100099. doi: 10.1016/j.bbadva.2023.100099. eCollection 2023. BBA Adv. 2023. PMID: 37675199 Free PMC article.
-
Arginine Residues Modulate the Membrane Interactions of pHLIP Peptides.J Chem Inf Model. 2023 Jul 24;63(14):4433-4446. doi: 10.1021/acs.jcim.3c00360. Epub 2023 Jul 3. J Chem Inf Model. 2023. PMID: 37395685 Free PMC article.
-
Targeting Lysosomes: A Strategy Against Chemoresistance in Cancer.Mini Rev Med Chem. 2024;24(15):1449-1468. doi: 10.2174/0113895575287242240129120002. Mini Rev Med Chem. 2024. PMID: 38343053 Review.
-
Amantadine: reappraisal of the timeless diamond-target updates and novel therapeutic potentials.J Neural Transm (Vienna). 2021 Feb;128(2):127-169. doi: 10.1007/s00702-021-02306-2. Epub 2021 Feb 23. J Neural Transm (Vienna). 2021. PMID: 33624170 Free PMC article. Review.
-
Getting Lost in the Cell-Lysosomal Entrapment of Chemotherapeutics.Cancers (Basel). 2020 Dec 7;12(12):3669. doi: 10.3390/cancers12123669. Cancers (Basel). 2020. PMID: 33297435 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

