Outlining the Complex Pathway of Mammalian Fe-S Cluster Biogenesis
- PMID: 32311335
- PMCID: PMC8349188
- DOI: 10.1016/j.tibs.2020.02.001
Outlining the Complex Pathway of Mammalian Fe-S Cluster Biogenesis
Abstract
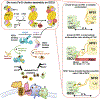
Iron-sulfur (Fe-S) clusters (ISCs) are ubiquitous cofactors essential to numerous fundamental cellular processes. Assembly of ISCs and their insertion into apoproteins involves the function of complex cellular machineries that operate in parallel in the mitochondrial and cytosolic/nuclear compartments of mammalian cells. The spectrum of diseases caused by inherited defects in genes that encode the Fe-S assembly proteins has recently expanded to include multiple rare human diseases, which manifest distinctive combinations and severities of global and tissue-specific impairments. In this review, we provide an overview of our understanding of ISC biogenesis in mammalian cells, discuss recent work that has shed light on the molecular interactions that govern ISC assembly, and focus on human diseases caused by failures of the biogenesis pathway.
Keywords: CIAO1; FAM96B; HSC20; HSPA9; MMS19; frataxin; mitochondrial iron overload; multiple mitochondrial dysfunctions syndromes; secondary carriers.
Published by Elsevier Ltd.
Figures

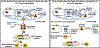
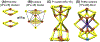
Similar articles
-
Cytosolic HSC20 integrates de novo iron-sulfur cluster biogenesis with the CIAO1-mediated transfer to recipients.Hum Mol Genet. 2018 Mar 1;27(5):837-852. doi: 10.1093/hmg/ddy004. Hum Mol Genet. 2018. PMID: 29309586 Free PMC article.
-
Iron-sulfur cluster biogenesis in mammalian cells: New insights into the molecular mechanisms of cluster delivery.Biochim Biophys Acta. 2015 Jun;1853(6):1493-512. doi: 10.1016/j.bbamcr.2014.09.009. Epub 2014 Sep 19. Biochim Biophys Acta. 2015. PMID: 25245479 Free PMC article. Review.
-
Mechanisms of iron-sulfur protein maturation in mitochondria, cytosol and nucleus of eukaryotes.Biochim Biophys Acta. 2006 Jul;1763(7):652-67. doi: 10.1016/j.bbamcr.2006.05.011. Epub 2006 May 23. Biochim Biophys Acta. 2006. PMID: 16843540 Review.
-
HSC20 interacts with frataxin and is involved in iron-sulfur cluster biogenesis and iron homeostasis.Hum Mol Genet. 2012 Apr 1;21(7):1457-69. doi: 10.1093/hmg/ddr582. Epub 2011 Dec 13. Hum Mol Genet. 2012. PMID: 22171070 Free PMC article.
-
The Toxoplasma gondii mitochondrial transporter ABCB7L is essential for the biogenesis of cytosolic and nuclear iron-sulfur cluster proteins and cytosolic translation.mBio. 2024 Oct 16;15(10):e0087224. doi: 10.1128/mbio.00872-24. Epub 2024 Aug 29. mBio. 2024. PMID: 39207139 Free PMC article.
Cited by
-
Fe-S cofactors in the SARS-CoV-2 RNA-dependent RNA polymerase are potential antiviral targets.Science. 2021 Jul 9;373(6551):236-241. doi: 10.1126/science.abi5224. Epub 2021 Jun 3. Science. 2021. PMID: 34083449 Free PMC article.
-
Effects of Three-Month Administration of High-Saturated Fat Diet and High-Polyunsaturated Fat Diets with Different Linoleic Acid (LA, C18:2n-6) to α-Linolenic Acid (ALA, C18:3n-3) Ratio on the Mouse Liver Proteome.Nutrients. 2021 May 15;13(5):1678. doi: 10.3390/nu13051678. Nutrients. 2021. PMID: 34063343 Free PMC article.
-
The Role of S-Glutathionylation in Health and Disease: A Bird's Eye View.Nutrients. 2024 Aug 18;16(16):2753. doi: 10.3390/nu16162753. Nutrients. 2024. PMID: 39203889 Free PMC article. Review.
-
Structure of a putative immature form of a Rieske-type iron-sulfur protein in complex with zinc chloride.Commun Chem. 2023 Sep 9;6(1):190. doi: 10.1038/s42004-023-01000-6. Commun Chem. 2023. PMID: 37689761 Free PMC article.
-
The long-standing relationship between paramagnetic NMR and iron-sulfur proteins: the mitoNEET example. An old method for new stories or the other way around?Magn Reson (Gott). 2021 Apr 26;2(1):203-221. doi: 10.5194/mr-2-203-2021. eCollection 2021. Magn Reson (Gott). 2021. PMID: 37904758 Free PMC article.
References
-
- Martin W et al. (2008) Hydrothermal vents and the origin of life. Nat. Rev. Microbiol 6, 805–814 - PubMed
-
- Beinert H et al. (1997) Iron-sulfur clusters: nature’s modular, multipurpose structures. Science 277, 653–659 - PubMed
-
- Beinert H et al. (1996) Aconitase as iron-sulfur protein, enzyme, and iron-regulatory protein. Chem. Rev 96, 2335–2374 - PubMed
-
- Dailey HA et al. (1994) Human ferrochelatase is an iron-sulfur protein. Biochemistry 33, 403–407 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

