The Catalytic-Dependent and -Independent Roles of Lsd1 and Lsd2 Lysine Demethylases in Heterochromatin Formation in Schizosaccharomyces pombe
- PMID: 32295063
- PMCID: PMC7226997
- DOI: 10.3390/cells9040955
The Catalytic-Dependent and -Independent Roles of Lsd1 and Lsd2 Lysine Demethylases in Heterochromatin Formation in Schizosaccharomyces pombe
Abstract
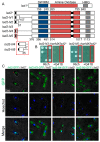
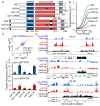
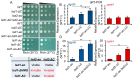
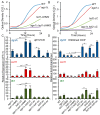
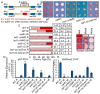
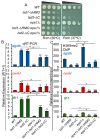
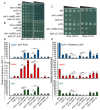
In eukaryotes, heterochromatin plays a critical role in organismal development and cell fate acquisition, through regulating gene expression. The evolutionarily conserved lysine-specific demethylases, Lsd1 and Lsd2, remove mono- and dimethylation on histone H3, serving complex roles in gene expression. In the fission yeast Schizosaccharomyces pombe, null mutations of Lsd1 and Lsd2 result in either severe growth defects or inviability, while catalytic inactivation causes minimal defects, indicating that Lsd1 and Lsd2 have essential functions beyond their known demethylase activity. Here, we show that catalytic mutants of Lsd1 or Lsd2 partially assemble functional heterochromatin at centromeres in RNAi-deficient cells, while the C-terminal truncated alleles of Lsd1 or Lsd2 exacerbate heterochromatin formation at all major heterochromatic regions, suggesting that Lsd1 and Lsd2 repress heterochromatic transcripts through mechanisms both dependent on and independent of their catalytic activities. Lsd1 and Lsd2 are also involved in the establishment and maintenance of heterochromatin. At constitutive heterochromatic regions, Lsd1 and Lsd2 regulate one another and cooperate with other histone modifiers, including the class II HDAC Clr3 and the Sirtuin family protein Sir2 for gene silencing, but not with the class I HDAC Clr6. Our findings explore the roles of lysine-specific demethylases in epigenetic gene silencing at heterochromatic regions.
Keywords: H3K9me2; Lsd1; Lsd2; gene silencing; heterochromatin; lysine demethylase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
The Cross-Regulation Between Set1, Clr4, and Lsd1/2 in Schizosaccharomyces pombe.PLoS Genet. 2024 Jan 5;20(1):e1011107. doi: 10.1371/journal.pgen.1011107. eCollection 2024 Jan. PLoS Genet. 2024. PMID: 38181050 Free PMC article.
-
LSD2 Is an Epigenetic Player in Multiple Types of Cancer and Beyond.Biomolecules. 2024 May 3;14(5):553. doi: 10.3390/biom14050553. Biomolecules. 2024. PMID: 38785960 Free PMC article. Review.
-
Regulation of tissue factor pathway inhibitor-2 (TFPI-2) expression by lysine-specific demethylase 1 and 2 (LSD1 and LSD2).Biosci Biotechnol Biochem. 2014;78(6):1010-7. doi: 10.1080/09168451.2014.910104. Epub 2014 Jun 13. Biosci Biotechnol Biochem. 2014. PMID: 25036127
-
Genome-wide studies of histone demethylation catalysed by the fission yeast homologues of mammalian LSD1.PLoS One. 2007 Apr 18;2(4):e386. doi: 10.1371/journal.pone.0000386. PLoS One. 2007. PMID: 17440621 Free PMC article.
-
The Role of LSD1 and LSD2 in Cancers of the Gastrointestinal System: An Update.Biomolecules. 2022 Mar 17;12(3):462. doi: 10.3390/biom12030462. Biomolecules. 2022. PMID: 35327654 Free PMC article. Review.
Cited by
-
The Cross-Regulation Between Set1, Clr4, and Lsd1/2 in Schizosaccharomyces pombe.PLoS Genet. 2024 Jan 5;20(1):e1011107. doi: 10.1371/journal.pgen.1011107. eCollection 2024 Jan. PLoS Genet. 2024. PMID: 38181050 Free PMC article.
-
Modified Histone Peptides Linked to Magnetic Beads Reduce Binding Specificity.Int J Mol Sci. 2022 Feb 1;23(3):1691. doi: 10.3390/ijms23031691. Int J Mol Sci. 2022. PMID: 35163614 Free PMC article.
-
To Erase or Not to Erase: Non-Canonical Catalytic Functions and Non-Catalytic Functions of Members of Histone Lysine Demethylase Families.Int J Mol Sci. 2024 Jun 24;25(13):6900. doi: 10.3390/ijms25136900. Int J Mol Sci. 2024. PMID: 39000010 Free PMC article. Review.
-
LSD2 Is an Epigenetic Player in Multiple Types of Cancer and Beyond.Biomolecules. 2024 May 3;14(5):553. doi: 10.3390/biom14050553. Biomolecules. 2024. PMID: 38785960 Free PMC article. Review.
-
The Fission Yeast Mating-Type Switching Motto: "One-for-Two" and "Two-for-One".Microbiol Mol Biol Rev. 2023 Mar 21;87(1):e0000821. doi: 10.1128/mmbr.00008-21. Epub 2023 Jan 11. Microbiol Mol Biol Rev. 2023. PMID: 36629411 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

