Calnexin cycle - structural features of the ER chaperone system
- PMID: 32285592
- PMCID: PMC7687155
- DOI: 10.1111/febs.15330
Calnexin cycle - structural features of the ER chaperone system
Abstract
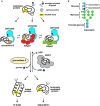
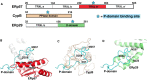
The endoplasmic reticulum (ER) is the major folding compartment for secreted and membrane proteins and is the site of a specific chaperone system, the calnexin cycle, for folding N-glycosylated proteins. Recent structures of components of the calnexin cycle have deepened our understanding of quality control mechanisms and protein folding pathways in the ER. In the calnexin cycle, proteins carrying monoglucosylated glycans bind to the lectin chaperones calnexin and calreticulin, which recruit a variety of function-specific chaperones to mediate protein disulfide formation, proline isomerization, and general protein folding. Upon trimming by glucosidase II, the glycan without an inner glucose residue is no longer able to bind to the lectin chaperones. For proteins that have not yet folded properly, the enzyme UDP-glucose:glycoprotein glucosyltransferase (UGGT) acts as a checkpoint by adding a glucose back to the N-glycan. This allows the misfolded proteins to re-associate with calnexin and calreticulin for additional rounds of chaperone-mediated refolding and prevents them from exiting the ERs. Here, we review progress in structural studies of the calnexin cycle, which reveal common features of how lectin chaperones recruit function-specific chaperones and how UGGT recognizes misfolded proteins.
Keywords: CypB; ERp29; ERp57; PDI; UGGT; calnexin; calnexin cycle; calreticulin; endoplasmic reticulum; protein folding.
© 2020 The Authors. The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum.J Cell Sci. 2006 Feb 15;119(Pt 4):615-23. doi: 10.1242/jcs.02856. J Cell Sci. 2006. PMID: 16467570 Review.
-
Mapping the ER Interactome: The P Domains of Calnexin and Calreticulin as Plurivalent Adapters for Foldases and Chaperones.Structure. 2017 Sep 5;25(9):1415-1422.e3. doi: 10.1016/j.str.2017.07.010. Structure. 2017. PMID: 28877505
-
Protein quality control in the ER: the recognition of misfolded proteins.Semin Cell Dev Biol. 2010 Jul;21(5):500-11. doi: 10.1016/j.semcdb.2010.03.006. Epub 2010 Mar 25. Semin Cell Dev Biol. 2010. PMID: 20347046 Review.
-
In vitro assays of the functions of calnexin and calreticulin, lectin chaperones of the endoplasmic reticulum.Methods Mol Biol. 2006;347:331-42. doi: 10.1385/1-59745-167-3:331. Methods Mol Biol. 2006. PMID: 17072021 Review.
-
Dimerization of ER-resident molecular chaperones mediated by ERp29.Biochem Biophys Res Commun. 2021 Jan 15;536:52-58. doi: 10.1016/j.bbrc.2020.12.023. Epub 2020 Dec 25. Biochem Biophys Res Commun. 2021. PMID: 33360823
Cited by
-
Influence of glycosylation on the immunogenicity and antigenicity of viral immunogens.Biotechnol Adv. 2024 Jan-Feb;70:108283. doi: 10.1016/j.biotechadv.2023.108283. Epub 2023 Nov 14. Biotechnol Adv. 2024. PMID: 37972669 Free PMC article. Review.
-
Targeting Endoplasmic Reticulum Stress by Natural and Chemical Compounds Ameliorates Cisplatin-Induced Nephrotoxicity: A Review.Biol Trace Elem Res. 2024 Aug 30. doi: 10.1007/s12011-024-04351-w. Online ahead of print. Biol Trace Elem Res. 2024. PMID: 39212819 Review.
-
Mixed mechanism of conformational selection and induced fit as a molecular recognition process in the calreticulin family of proteins.PLoS Comput Biol. 2022 Dec 12;18(12):e1010661. doi: 10.1371/journal.pcbi.1010661. eCollection 2022 Dec. PLoS Comput Biol. 2022. PMID: 36508460 Free PMC article.
-
Comprehensive proteomics and meta-analysis of COVID-19 host response.Nat Commun. 2023 Sep 22;14(1):5921. doi: 10.1038/s41467-023-41159-z. Nat Commun. 2023. PMID: 37739942 Free PMC article.
-
The T-type calcium channelosome.Pflugers Arch. 2024 Feb;476(2):163-177. doi: 10.1007/s00424-023-02891-z. Epub 2023 Dec 1. Pflugers Arch. 2024. PMID: 38036777 Review.
References
-
- Ellgaard L, McCaul N, Chatsisvili A & Braakman I (2016) Co‐ and post‐translational protein folding in the ER. Traffic 17, 615–638. - PubMed
-
- Kozlov G, Maattanen P, Thomas DY & Gehring K (2010) A structural overview of the PDI family of proteins. FEBS J 277, 3924–3936. - PubMed
-
- Hatahet F & Ruddock LW (2009) Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal 11, 2807–2850. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

