GOMCL: a toolkit to cluster, evaluate, and extract non-redundant associations of Gene Ontology-based functions
- PMID: 32272889
- PMCID: PMC7146957
- DOI: 10.1186/s12859-020-3447-4
GOMCL: a toolkit to cluster, evaluate, and extract non-redundant associations of Gene Ontology-based functions
Abstract
Background: Functional enrichment of genes and pathways based on Gene Ontology (GO) has been widely used to describe the results of various -omics analyses. GO terms statistically overrepresented within a set of a large number of genes are typically used to describe the main functional attributes of the gene set. However, these lists of overrepresented GO terms are often too large and contains redundant overlapping GO terms hindering informative functional interpretations.
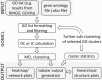
Results: We developed GOMCL to reduce redundancy and summarize lists of GO terms effectively and informatively. This lightweight python toolkit efficiently identifies clusters within a list of GO terms using the Markov Clustering (MCL) algorithm, based on the overlap of gene members between GO terms. GOMCL facilitates biological interpretation of a large number of GO terms by condensing them into GO clusters representing non-overlapping functional themes. It enables visualizing GO clusters as a heatmap, networks based on either overlap of members or hierarchy among GO terms, and tables with depth and cluster information for each GO term. Each GO cluster generated by GOMCL can be evaluated and further divided into non-overlapping sub-clusters using the GOMCL-sub module. The outputs from both GOMCL and GOMCL-sub can be imported to Cytoscape for additional visualization effects.
Conclusions: GOMCL is a convenient toolkit to cluster, evaluate, and extract non-redundant associations of Gene Ontology-based functions. GOMCL helps researchers to reduce time spent on manual curation of large lists of GO terms, minimize biases introduced by redundant GO terms in data interpretation, and batch processing of multiple GO enrichment datasets. A user guide, a test dataset, and the source code of GOMCL are available at https://github.com/Guannan-Wang/GOMCL and www.lsugenomics.org.
Keywords: Functional genomics; Functional networks; GO annotations; GO similarity; Gene ontology clustering; High throughput omics; Markov clustering.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Gene Ontology analysis in multiple gene clusters under multiple hypothesis testing framework.Artif Intell Med. 2007 Oct;41(2):105-15. doi: 10.1016/j.artmed.2007.08.002. Artif Intell Med. 2007. PMID: 17913480
-
GOTrapper: a tool to navigate through branches of gene ontology hierarchy.BMC Bioinformatics. 2019 Jan 11;20(1):20. doi: 10.1186/s12859-018-2581-8. BMC Bioinformatics. 2019. PMID: 30634902 Free PMC article.
-
NaviGO: interactive tool for visualization and functional similarity and coherence analysis with gene ontology.BMC Bioinformatics. 2017 Mar 20;18(1):177. doi: 10.1186/s12859-017-1600-5. BMC Bioinformatics. 2017. PMID: 28320317 Free PMC article.
-
Black sheep, dark horses, and colorful dogs: a review on the current state of the Gene Ontology with respect to iron homeostasis in Arabidopsis thaliana.Front Plant Sci. 2023 Jul 24;14:1204723. doi: 10.3389/fpls.2023.1204723. eCollection 2023. Front Plant Sci. 2023. PMID: 37554559 Free PMC article. Review.
-
DiVenn: An Interactive and Integrated Web-Based Visualization Tool for Comparing Gene Lists.Front Genet. 2019 May 3;10:421. doi: 10.3389/fgene.2019.00421. eCollection 2019. Front Genet. 2019. PMID: 31130993 Free PMC article. Review.
Cited by
-
Living with high potassium: Balance between nutrient acquisition and K-induced salt stress signaling.Plant Physiol. 2023 Feb 12;191(2):1102-1121. doi: 10.1093/plphys/kiac564. Plant Physiol. 2023. PMID: 36493387 Free PMC article.
-
Novel genome characteristics contribute to the invasiveness of Phragmites australis (common reed).Mol Ecol. 2022 Feb;31(4):1142-1159. doi: 10.1111/mec.16293. Epub 2021 Dec 11. Mol Ecol. 2022. PMID: 34839548 Free PMC article.
-
Transcription and DNA methylation signatures of paternal behavior in hippocampal dentate gyrus of prairie voles.Sci Rep. 2023 Jul 7;13(1):11020. doi: 10.1038/s41598-023-37521-2. Sci Rep. 2023. PMID: 37419920 Free PMC article.
-
vissE: a versatile tool to identify and visualise higher-order molecular phenotypes from functional enrichment analysis.BMC Bioinformatics. 2024 Feb 8;25(1):64. doi: 10.1186/s12859-024-05676-y. BMC Bioinformatics. 2024. PMID: 38331751 Free PMC article.
-
Genes Associated with Biological Nitrogen Fixation Efficiency Identified Using RNA Sequencing in Red Clover (Trifolium pratense L.).Life (Basel). 2022 Nov 25;12(12):1975. doi: 10.3390/life12121975. Life (Basel). 2022. PMID: 36556339 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

