Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing
- PMID: 32251383
- PMCID: PMC7735425
- DOI: 10.1038/s41565-020-0669-6
Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing
Abstract
CRISPR-Cas gene editing and messenger RNA-based protein replacement therapy hold tremendous potential to effectively treat disease-causing mutations with diverse cellular origin. However, it is currently impossible to rationally design nanoparticles that selectively target specific tissues. Here, we report a strategy termed selective organ targeting (SORT) wherein multiple classes of lipid nanoparticles are systematically engineered to exclusively edit extrahepatic tissues via addition of a supplemental SORT molecule. Lung-, spleen- and liver-targeted SORT lipid nanoparticles were designed to selectively edit therapeutically relevant cell types including epithelial cells, endothelial cells, B cells, T cells and hepatocytes. SORT is compatible with multiple gene editing techniques, including mRNA, Cas9 mRNA/single guide RNA and Cas9 ribonucleoprotein complexes, and is envisioned to aid the development of protein replacement and gene correction therapeutics in targeted tissues.
Conflict of interest statement
Competing interests
D.J.S., Q.C., T.W., and the Reagents of the University of Texas System have filed patent applications on SORT and related technologies. D.J.S. is a co-founder of ReCode Therapeutics, which has licensed intellectual property from UT Southwestern.
Figures


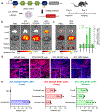
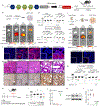
Comment in
-
Nanotechnology for organ-tunable gene editing.Nat Nanotechnol. 2020 Apr;15(4):253-255. doi: 10.1038/s41565-020-0666-9. Nat Nanotechnol. 2020. PMID: 32251384 No abstract available.
Similar articles
-
Cell-Selective Messenger RNA Delivery and CRISPR/Cas9 Genome Editing by Modulating the Interface of Phenylboronic Acid-Derived Lipid Nanoparticles and Cellular Surface Sialic Acid.ACS Appl Mater Interfaces. 2019 Dec 18;11(50):46585-46590. doi: 10.1021/acsami.9b17749. Epub 2019 Dec 9. ACS Appl Mater Interfaces. 2019. PMID: 31763806
-
Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR-Cas gene editing.Nat Mater. 2021 May;20(5):701-710. doi: 10.1038/s41563-020-00886-0. Epub 2021 Feb 4. Nat Mater. 2021. PMID: 33542471 Free PMC article.
-
Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3.Proc Natl Acad Sci U S A. 2021 Mar 9;118(10):e2020401118. doi: 10.1073/pnas.2020401118. Proc Natl Acad Sci U S A. 2021. PMID: 33649229 Free PMC article.
-
Intracellular Delivery of mRNA for Cell-Selective CRISPR/Cas9 Genome Editing using Lipid Nanoparticles.Chembiochem. 2023 May 2;24(9):e202200801. doi: 10.1002/cbic.202200801. Epub 2023 Mar 30. Chembiochem. 2023. PMID: 36780174 Review.
-
Preparation of selective organ-targeting (SORT) lipid nanoparticles (LNPs) using multiple technical methods for tissue-specific mRNA delivery.Nat Protoc. 2023 Jan;18(1):265-291. doi: 10.1038/s41596-022-00755-x. Epub 2022 Oct 31. Nat Protoc. 2023. PMID: 36316378 Free PMC article. Review.
Cited by
-
Membrane-destabilizing ionizable phospholipids: Novel components for organ-selective mRNA delivery and CRISPR-Cas gene editing.Signal Transduct Target Ther. 2021 May 25;6(1):206. doi: 10.1038/s41392-021-00642-z. Signal Transduct Target Ther. 2021. PMID: 34035211 Free PMC article. No abstract available.
-
Engineering Lipid Nanoparticles for Enhanced Intracellular Delivery of mRNA through Inhalation.ACS Nano. 2022 Sep 27;16(9):14792-14806. doi: 10.1021/acsnano.2c05647. Epub 2022 Aug 29. ACS Nano. 2022. PMID: 36038136 Free PMC article.
-
Lipid Nanoparticle (LNP) Chemistry Can Endow Unique In Vivo RNA Delivery Fates within the Liver That Alter Therapeutic Outcomes in a Cancer Model.Mol Pharm. 2022 Nov 7;19(11):3973-3986. doi: 10.1021/acs.molpharmaceut.2c00442. Epub 2022 Sep 26. Mol Pharm. 2022. PMID: 36154076 Free PMC article.
-
Lipid nanovehicles overcome barriers to systemic RNA delivery: Lipid components, fabrication methods, and rational design.Acta Pharm Sin B. 2024 Feb;14(2):579-601. doi: 10.1016/j.apsb.2023.10.012. Epub 2023 Oct 20. Acta Pharm Sin B. 2024. PMID: 38322344 Free PMC article. Review.
-
Novel Strategy to Combat Antibiotic Resistance: A Sight into the Combination of CRISPR/Cas9 and Nanoparticles.Pharmaceutics. 2021 Mar 8;13(3):352. doi: 10.3390/pharmaceutics13030352. Pharmaceutics. 2021. PMID: 33800235 Free PMC article. Review.
References
-
- Doudna JA & Charpentier E Genome editing: The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 (2014). - PubMed
-
- Wang HX et al. CRISPR/Cas9-based genome editing for disease modeling and therapy: Challenges and opportunities for nonviral delivery. Chem. Rev 117, 9874–9906 (2017). - PubMed
-
- Hajj KA & Whitehead KA Tools for translation: Non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater 2, 17056 (2017).
-
- Sahin U, Kariko K & Tureci O mRNA-based therapeutics: Developing a new class of drugs. Nat. Rev. Drug Discovery 13, 759–780 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

