Structural basis of receptor recognition by SARS-CoV-2
- PMID: 32225175
- PMCID: PMC7328981
- DOI: 10.1038/s41586-020-2179-y
Structural basis of receptor recognition by SARS-CoV-2
Abstract
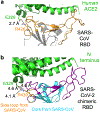
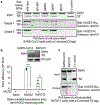
A novel severe acute respiratory syndrome (SARS)-like coronavirus (SARS-CoV-2) recently emerged and is rapidly spreading in humans, causing COVID-191,2. A key to tackling this pandemic is to understand the receptor recognition mechanism of the virus, which regulates its infectivity, pathogenesis and host range. SARS-CoV-2 and SARS-CoV recognize the same receptor-angiotensin-converting enzyme 2 (ACE2)-in humans3,4. Here we determined the crystal structure of the receptor-binding domain (RBD) of the spike protein of SARS-CoV-2 (engineered to facilitate crystallization) in complex with ACE2. In comparison with the SARS-CoV RBD, an ACE2-binding ridge in SARS-CoV-2 RBD has a more compact conformation; moreover, several residue changes in the SARS-CoV-2 RBD stabilize two virus-binding hotspots at the RBD-ACE2 interface. These structural features of SARS-CoV-2 RBD increase its ACE2-binding affinity. Additionally, we show that RaTG13, a bat coronavirus that is closely related to SARS-CoV-2, also uses human ACE2 as its receptor. The differences among SARS-CoV-2, SARS-CoV and RaTG13 in ACE2 recognition shed light on the potential animal-to-human transmission of SARS-CoV-2. This study provides guidance for intervention strategies that target receptor recognition by SARS-CoV-2.
Figures



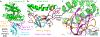
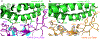
Similar articles
-
Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2.J Med Virol. 2020 Jun;92(6):595-601. doi: 10.1002/jmv.25726. Epub 2020 Mar 11. J Med Virol. 2020. PMID: 32100877 Free PMC article.
-
Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus.J Virol. 2020 Mar 17;94(7):e00127-20. doi: 10.1128/JVI.00127-20. Print 2020 Mar 17. J Virol. 2020. PMID: 31996437 Free PMC article.
-
Role of the GTNGTKR motif in the N-terminal receptor-binding domain of the SARS-CoV-2 spike protein.Virus Res. 2020 Sep;286:198058. doi: 10.1016/j.virusres.2020.198058. Epub 2020 Jun 9. Virus Res. 2020. PMID: 32531235 Free PMC article.
-
Mass Spectrometry and Structural Biology Techniques in the Studies on the Coronavirus-Receptor Interaction.Molecules. 2020 Sep 10;25(18):4133. doi: 10.3390/molecules25184133. Molecules. 2020. PMID: 32927621 Free PMC article. Review.
-
Unraveling the Epidemiology, Geographical Distribution, and Genomic Evolution of Potentially Lethal Coronaviruses (SARS, MERS, and SARS CoV-2).Front Cell Infect Microbiol. 2020 Aug 27;10:499. doi: 10.3389/fcimb.2020.00499. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32974224 Free PMC article. Review.
Cited by
-
S-RBD-modified and miR-486-5p-engineered exosomes derived from mesenchymal stem cells suppress ferroptosis and alleviate radiation-induced lung injury and long-term pulmonary fibrosis.J Nanobiotechnology. 2024 Oct 26;22(1):662. doi: 10.1186/s12951-024-02830-9. J Nanobiotechnology. 2024. PMID: 39462403 Free PMC article.
-
An Analysis of Combined Molecular Weight and Hydrophobicity Similarity between the Amino Acid Sequences of Spike Protein Receptor Binding Domains of Betacoronaviruses and Functionally Similar Sequences from Other Virus Families.Microorganisms. 2024 Oct 5;12(10):2021. doi: 10.3390/microorganisms12102021. Microorganisms. 2024. PMID: 39458330 Free PMC article.
-
Peptide-Based Inhibitors of Protein-Protein Interactions (PPIs): A Case Study on the Interaction Between SARS-CoV-2 Spike Protein and Human Angiotensin-Converting Enzyme 2 (hACE2).Biomedicines. 2024 Oct 16;12(10):2361. doi: 10.3390/biomedicines12102361. Biomedicines. 2024. PMID: 39457672 Free PMC article. Review.
-
Pulling Forces Differentially Affect Refolding Pathways Due to Entangled Misfolded States in SARS-CoV-1 and SARS-CoV-2 Receptor Binding Domain.Biomolecules. 2024 Oct 18;14(10):1327. doi: 10.3390/biom14101327. Biomolecules. 2024. PMID: 39456260 Free PMC article.
-
Evaluation of Angiotensin-Converting Enzyme 2 Expression In Vivo with Novel 68Ga-Labeled Peptides Originated from the Coronavirus Receptor-Binding Domain.ACS Pharmacol Transl Sci. 2024 Sep 12;7(10):3119-3130. doi: 10.1021/acsptsci.4c00316. eCollection 2024 Oct 11. ACS Pharmacol Transl Sci. 2024. PMID: 39416971
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

