Knockout of the OsNAC006 Transcription Factor Causes Drought and Heat Sensitivity in Rice
- PMID: 32225072
- PMCID: PMC7177362
- DOI: 10.3390/ijms21072288
Knockout of the OsNAC006 Transcription Factor Causes Drought and Heat Sensitivity in Rice
Abstract
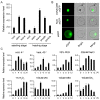
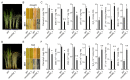
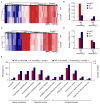
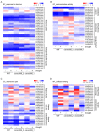
Rice (Oryza sativa) responds to various abiotic stresses during growth. Plant-specific NAM, ATAF1/2, and CUC2 (NAC) transcription factors (TFs) play an important role in controlling numerous vital growth and developmental processes. To date, 170 NAC TFs have been reported in rice, but their roles remain largely unknown. Herein, we discovered that the TF OsNAC006 is constitutively expressed in rice, and regulated by H2O2, cold, heat, abscisic acid (ABA), indole-3-acetic acid (IAA), gibberellin (GA), NaCl, and polyethylene glycol (PEG) 6000 treatments. Furthermore, knockout of OsNAC006 using the CRISPR-Cas9 system resulted in drought and heat sensitivity. RNA sequencing (RNA-seq) transcriptome analysis revealed that OsNAC006 regulates the expression of genes mainly involved in response to stimuli, oxidoreductase activity, cofactor binding, and membrane-related pathways. Our findings elucidate the important role of OsNAC006 in drought responses, and provide valuable information for genetic manipulation to enhance stress tolerance in future plant breeding programs.
Keywords: CRISPR-Cas9; NAC transcription factor; abiotic stresses; rice; transcriptome analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance.BMC Plant Biol. 2016 Sep 20;16(1):203. doi: 10.1186/s12870-016-0897-y. BMC Plant Biol. 2016. PMID: 27646344 Free PMC article.
-
Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response.BMC Plant Biol. 2019 Jun 25;19(1):278. doi: 10.1186/s12870-019-1883-y. BMC Plant Biol. 2019. PMID: 31238869 Free PMC article.
-
A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice.J Exp Bot. 2015 Nov;66(21):6803-17. doi: 10.1093/jxb/erv386. Epub 2015 Aug 10. J Exp Bot. 2015. PMID: 26261267 Free PMC article.
-
Modulating rice stress tolerance by transcription factors.Biotechnol Genet Eng Rev. 2008;25:381-403. doi: 10.5661/bger-25-381. Biotechnol Genet Eng Rev. 2008. PMID: 21412363 Review.
-
NAC Transcription Factors as Positive or Negative Regulators during Ongoing Battle between Pathogens and Our Food Crops.Int J Mol Sci. 2020 Dec 23;22(1):81. doi: 10.3390/ijms22010081. Int J Mol Sci. 2020. PMID: 33374758 Free PMC article. Review.
Cited by
-
Rapid breeding of an early maturing, high-quality, and high-y.ielding rice cultivar using marker‑assisted selection coupled with optimized anther culture.Mol Breed. 2024 Sep 6;44(9):58. doi: 10.1007/s11032-024-01495-4. eCollection 2024 Sep. Mol Breed. 2024. PMID: 39246623
-
Genome Editing and Improvement of Abiotic Stress Tolerance in Crop Plants.Life (Basel). 2023 Jun 27;13(7):1456. doi: 10.3390/life13071456. Life (Basel). 2023. PMID: 37511831 Free PMC article. Review.
-
Comprehensive Analysis of NAC Genes Reveals Differential Expression Patterns in Response to Pst DC3000 and Their Overlapping Expression Pattern during PTI and ETI in Tomato.Genes (Basel). 2022 Nov 2;13(11):2015. doi: 10.3390/genes13112015. Genes (Basel). 2022. PMID: 36360252 Free PMC article. Review.
-
Gene Editing for Plant Resistance to Abiotic Factors: A Systematic Review.Plants (Basel). 2023 Jan 9;12(2):305. doi: 10.3390/plants12020305. Plants (Basel). 2023. PMID: 36679018 Free PMC article. Review.
-
Overexpression of TaSNAC4-3D in Common Wheat (Triticum aestivum L.) Negatively Regulates Drought Tolerance.Front Plant Sci. 2022 Jul 4;13:945272. doi: 10.3389/fpls.2022.945272. eCollection 2022. Front Plant Sci. 2022. PMID: 35860542 Free PMC article.
References
-
- Nguyen H.M., Sako K., Matsui A., Suzuki Y., Mostofa M.G., Ha C.V., Tanaka M., Tran L.P., Habu Y., Seki M. Ethanol Enhances High-Salinity Stress Tolerance by Detoxifying Reactive Oxygen Species in Arabidopsis thaliana and Rice. Front. Plant Sci. 2017;8:1001. doi: 10.3389/fpls.2017.01001. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

