Phase separation directs ubiquitination of gene-body nucleosomes
- PMID: 32214243
- PMCID: PMC7481934
- DOI: 10.1038/s41586-020-2097-z
Phase separation directs ubiquitination of gene-body nucleosomes
Abstract
The conserved yeast E3 ubiquitin ligase Bre1 and its partner, the E2 ubiquitin-conjugating enzyme Rad6, monoubiquitinate histone H2B across gene bodies during the transcription cycle1. Although processive ubiquitination might-in principle-arise from Bre1 and Rad6 travelling with RNA polymerase II2, the mechanism of H2B ubiquitination across genic nucleosomes remains unclear. Here we implicate liquid-liquid phase separation3 as the underlying mechanism. Biochemical reconstitution shows that Bre1 binds the scaffold protein Lge1, which possesses an intrinsically disordered region that phase-separates via multivalent interactions. The resulting condensates comprise a core of Lge1 encapsulated by an outer catalytic shell of Bre1. This layered liquid recruits Rad6 and the nucleosomal substrate, which accelerates the ubiquitination of H2B. In vivo, the condensate-forming region of Lge1 is required to ubiquitinate H2B in gene bodies beyond the +1 nucleosome. Our data suggest that layered condensates of histone-modifying enzymes generate chromatin-associated 'reaction chambers', with augmented catalytic activity along gene bodies. Equivalent processes may occur in human cells, and cause neurological disease when impaired.
Conflict of interest statement
Declaration of interests
B.F.P. has a financial interest in Peconic, LLC, which utilizes the ChIP-exo technology implemented in this study and could potentially benefit from the outcomes of this research The remaining authors declare no competing interests.
Figures



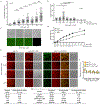
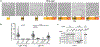



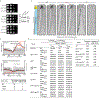
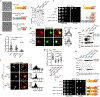



Comment in
-
Chromatin modified in a molecular reaction chamber.Nature. 2020 Mar;579(7800):503-504. doi: 10.1038/d41586-020-00638-9. Nature. 2020. PMID: 32161343 No abstract available.
-
Liquid phase condensation directs nucleosome epigenetic modifications.Signal Transduct Target Ther. 2020 May 6;5(1):64. doi: 10.1038/s41392-020-0166-2. Signal Transduct Target Ther. 2020. PMID: 32376821 Free PMC article. No abstract available.
Similar articles
-
Monoubiquitination of histone H2B is intrinsic to the Bre1 RING domain-Rad6 interaction and augmented by a second Rad6-binding site on Bre1.J Biol Chem. 2015 Feb 27;290(9):5298-310. doi: 10.1074/jbc.M114.626788. Epub 2014 Dec 29. J Biol Chem. 2015. PMID: 25548288 Free PMC article.
-
The Bre1/Rad6 machinery: writing the central histone ubiquitin mark on H2B and beyond.Chromosome Res. 2020 Dec;28(3-4):247-258. doi: 10.1007/s10577-020-09640-3. Epub 2020 Sep 7. Chromosome Res. 2020. PMID: 32895784 Review.
-
Structural mechanism for the recognition and ubiquitination of a single nucleosome residue by Rad6-Bre1.Proc Natl Acad Sci U S A. 2016 Sep 20;113(38):10553-8. doi: 10.1073/pnas.1606863113. Epub 2016 Sep 6. Proc Natl Acad Sci U S A. 2016. PMID: 27601672 Free PMC article.
-
Bre1 mediates the ubiquitination of histone H2B by regulating Lge1 stability.FEBS Lett. 2018 May;592(9):1565-1574. doi: 10.1002/1873-3468.13049. Epub 2018 Apr 24. FEBS Lett. 2018. PMID: 29637554
-
Histone H2B ubiquitylation: Connections to transcription and effects on chromatin structure.Biochim Biophys Acta Gene Regul Mech. 2024 Jun;1867(2):195018. doi: 10.1016/j.bbagrm.2024.195018. Epub 2024 Feb 6. Biochim Biophys Acta Gene Regul Mech. 2024. PMID: 38331024 Review.
Cited by
-
Ubiquitin-dependent regulation of transcription in development and disease.EMBO Rep. 2021 Apr 7;22(4):e51078. doi: 10.15252/embr.202051078. Epub 2021 Mar 28. EMBO Rep. 2021. PMID: 33779035 Free PMC article. Review.
-
Single-molecule imaging of epigenetic complexes in living cells: insights from studies on Polycomb group proteins.Nucleic Acids Res. 2021 Jul 9;49(12):6621-6637. doi: 10.1093/nar/gkab304. Nucleic Acids Res. 2021. PMID: 34009336 Free PMC article. Review.
-
Histone post-translational modifications - cause and consequence of genome function.Nat Rev Genet. 2022 Sep;23(9):563-580. doi: 10.1038/s41576-022-00468-7. Epub 2022 Mar 25. Nat Rev Genet. 2022. PMID: 35338361 Review.
-
Control of protein stability by post-translational modifications.Nat Commun. 2023 Jan 13;14(1):201. doi: 10.1038/s41467-023-35795-8. Nat Commun. 2023. PMID: 36639369 Free PMC article. Review.
-
Feedback regulation of ubiquitination and phase separation of HECT E3 ligases.Proc Natl Acad Sci U S A. 2023 Aug 15;120(33):e2302478120. doi: 10.1073/pnas.2302478120. Epub 2023 Aug 7. Proc Natl Acad Sci U S A. 2023. PMID: 37549262 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

