Unrevealing sequence and structural features of novel coronavirus using in silico approaches: The main protease as molecular target
- PMID: 32210741
- PMCID: PMC7081067
- DOI: 10.17179/excli2020-1189
Unrevealing sequence and structural features of novel coronavirus using in silico approaches: The main protease as molecular target
Abstract
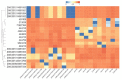
Direct-acting antivirals are effective tools to control viral infections. SARS-CoV-2 is a coronavirus associated with the epidemiological outbreak in late 2019. Previous reports showed that HIV-1 protease inhibitors could block SARS-CoV main protease. Based on that and using an in silico approach, we evaluated SARS-CoV-2 main protease as a target for HIV-1 protease inhibitors to reveal the structural features related to their antiviral effect. Our results showed that several HIV inhibitors such as lopinavir, ritonavir, and saquinavir produce strong interaction with the active site of SARS-CoV-2 main protease. Furthermore, broad library protease inhibitors obtained from PubChem and ZINC (www.zinc.docking.org) were evaluated. Our analysis revealed 20 compounds that could be clustered into three groups based on their chemical features. Then, these structures could serve as leading compounds to develop a series of derivatives optimizing their activity against SARS-CoV-2 and other coronaviruses. Altogether, the results presented in this work contribute to gain a deep understanding of the molecular pharmacology of SARS-CoV-2 treatment and validate the use of protease inhibitors against SARS-CoV-2.
Keywords: Coronavirus; HIV; SARS-CoV-2; protease; treatment.
Copyright © 2020 Ortega et al.
Figures


Similar articles
-
Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug.Am J Cancer Res. 2020 Aug 1;10(8):2535-2545. eCollection 2020. Am J Cancer Res. 2020. PMID: 32905393 Free PMC article.
-
Computational Evaluation of the Inhibition Efficacies of HIV Antivirals on SARS-CoV-2 (COVID-19) Protease and Identification of 3D Pharmacophore and Hit Compounds.Adv Pharmacol Pharm Sci. 2020 Sep 21;2020:8818008. doi: 10.1155/2020/8818008. eCollection 2020. Adv Pharmacol Pharm Sci. 2020. PMID: 33015628 Free PMC article.
-
Quinolines-Based SARS-CoV-2 3CLpro and RdRp Inhibitors and Spike-RBD-ACE2 Inhibitor for Drug-Repurposing Against COVID-19: An in silico Analysis.Front Microbiol. 2020 Jul 23;11:1796. doi: 10.3389/fmicb.2020.01796. eCollection 2020. Front Microbiol. 2020. PMID: 32793181 Free PMC article.
-
Virtual screening for functional foods against the main protease of SARS-CoV-2.J Food Biochem. 2020 Nov;44(11):e13481. doi: 10.1111/jfbc.13481. Epub 2020 Sep 27. J Food Biochem. 2020. PMID: 32984999
-
In-Silico Approaches for the Screening and Discovery of Broad-Spectrum Marine Natural Product Antiviral Agents Against Coronaviruses.Infect Drug Resist. 2023 Apr 19;16:2321-2338. doi: 10.2147/IDR.S395203. eCollection 2023. Infect Drug Resist. 2023. PMID: 37155475 Free PMC article. Review.
Cited by
-
Understanding Severe Acute Respiratory Syndrome Coronavirus 2 Replication to Design Efficient Drug Combination Therapies.Intervirology. 2020;63(1-6):2-9. doi: 10.1159/000512141. Epub 2020 Oct 23. Intervirology. 2020. PMID: 33099545 Free PMC article. Review.
-
Omicron SARS-CoV-2 Variant Spike Protein Shows an Increased Affinity to the Human ACE2 Receptor: An In Silico Analysis.Pathogens. 2021 Dec 31;11(1):45. doi: 10.3390/pathogens11010045. Pathogens. 2021. PMID: 35055993 Free PMC article.
-
Design, Synthesis, Molecular Modeling, Anticancer Studies, and Density Functional Theory Calculations of 4-(1,2,4-Triazol-3-ylsulfanylmethyl)-1,2,3-triazole Derivatives.ACS Omega. 2020 Dec 31;6(1):301-316. doi: 10.1021/acsomega.0c04595. eCollection 2021 Jan 12. ACS Omega. 2020. PMID: 33458482 Free PMC article.
-
Contributions of Latin American researchers in the understanding of the novel coronavirus outbreak: a literature review.PeerJ. 2020 Jun 4;8:e9332. doi: 10.7717/peerj.9332. eCollection 2020. PeerJ. 2020. PMID: 32547890 Free PMC article.
-
Class A G Protein-Coupled Receptor Antagonist Famotidine as a Therapeutic Alternative Against SARS-CoV2: An In Silico Analysis.Biomolecules. 2020 Jun 24;10(6):954. doi: 10.3390/biom10060954. Biomolecules. 2020. PMID: 32599963 Free PMC article.
References
-
- de Wilde AH, Jochmans D, Posthuma CC, Zevenhoven-Dobbe JC, van Nieuwkoop S, Bestebroer TM, et al. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob Agents Chemother. 2014;58:4875–84. - PMC - PubMed
-
- Khalid M, Al Rabiah F, Khan B, Al Mobeireek A, Butt TS, Al Mutairy E. Ribavirin and interferon-α2b as primary and preventive treatment for Middle East respiratory syndrome coronavirus: a preliminary report of two cases. Antivir Ther. 2015;20:87–91. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
