Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak
- PMID: 32197085
- PMCID: PMC7156161
- DOI: 10.1016/j.cub.2020.03.022
Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak
Erratum in
-
Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak.Curr Biol. 2020 Apr 20;30(8):1578. doi: 10.1016/j.cub.2020.03.063. Curr Biol. 2020. PMID: 32315626 Free PMC article. No abstract available.
Abstract
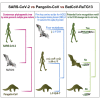
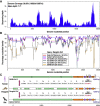
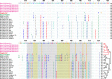
An outbreak of coronavirus disease 2019 (COVID-19) caused by the 2019 novel coronavirus (SARS-CoV-2) began in the city of Wuhan in China and has widely spread worldwide. Currently, it is vital to explore potential intermediate hosts of SARS-CoV-2 to control COVID-19 spread. Therefore, we reinvestigated published data from pangolin lung samples from which SARS-CoV-like CoVs were detected by Liu et al. [1]. We found genomic and evolutionary evidence of the occurrence of a SARS-CoV-2-like CoV (named Pangolin-CoV) in dead Malayan pangolins. Pangolin-CoV is 91.02% and 90.55% identical to SARS-CoV-2 and BatCoV RaTG13, respectively, at the whole-genome level. Aside from RaTG13, Pangolin-CoV is the most closely related CoV to SARS-CoV-2. The S1 protein of Pangolin-CoV is much more closely related to SARS-CoV-2 than to RaTG13. Five key amino acid residues involved in the interaction with human ACE2 are completely consistent between Pangolin-CoV and SARS-CoV-2, but four amino acid mutations are present in RaTG13. Both Pangolin-CoV and RaTG13 lost the putative furin recognition sequence motif at S1/S2 cleavage site that can be observed in the SARS-CoV-2. Conclusively, this study suggests that pangolin species are a natural reservoir of SARS-CoV-2-like CoVs.
Keywords: COVID-19; SARS-CoV-2; origin; pangolin.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures


Similar articles
-
Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins.Nature. 2020 Jul;583(7815):286-289. doi: 10.1038/s41586-020-2313-x. Epub 2020 May 7. Nature. 2020. PMID: 32380510
-
Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins.Nature. 2020 Jul;583(7815):282-285. doi: 10.1038/s41586-020-2169-0. Epub 2020 Mar 26. Nature. 2020. PMID: 32218527
-
COVID-19: Time to exonerate the pangolin from the transmission of SARS-CoV-2 to humans.Infect Genet Evol. 2020 Oct;84:104493. doi: 10.1016/j.meegid.2020.104493. Epub 2020 Aug 5. Infect Genet Evol. 2020. PMID: 32768565 Free PMC article.
-
Angiotensin-converting enzyme 2: The old door for new severe acute respiratory syndrome coronavirus 2 infection.Rev Med Virol. 2020 Sep;30(5):e2122. doi: 10.1002/rmv.2122. Epub 2020 Jun 30. Rev Med Virol. 2020. PMID: 32602627 Free PMC article. Review.
-
SARS-CoV-2, the pandemic coronavirus: Molecular and structural insights.J Basic Microbiol. 2021 Mar;61(3):180-202. doi: 10.1002/jobm.202000537. Epub 2021 Jan 18. J Basic Microbiol. 2021. PMID: 33460172 Free PMC article. Review.
Cited by
-
Detection of Mycotoxin Contamination in Foods Using Artificial Intelligence: A Review.Foods. 2024 Oct 21;13(20):3339. doi: 10.3390/foods13203339. Foods. 2024. PMID: 39456400 Free PMC article. Review.
-
On the Origin of SARS-CoV-2: Did Cell Culture Experiments Lead to Increased Virulence of the Progenitor Virus for Humans?In Vivo. 2021 May-Jun;35(3):1313-1326. doi: 10.21873/invivo.12384. Epub 2021 Apr 28. In Vivo. 2021. PMID: 33910809 Free PMC article. Review.
-
Isolation and characterization of a pangolin-borne HKU4-related coronavirus that potentially infects human-DPP4-transgenic mice.Nat Commun. 2024 Feb 5;15(1):1048. doi: 10.1038/s41467-024-45453-2. Nat Commun. 2024. PMID: 38316817 Free PMC article.
-
Rapid genomic characterization of SARS-CoV-2 viruses from clinical specimens using nanopore sequencing.Sci Rep. 2020 Oct 15;10(1):17492. doi: 10.1038/s41598-020-74656-y. Sci Rep. 2020. PMID: 33060796 Free PMC article.
-
COVID-19: pathogenesis, advances in treatment and vaccine development and environmental impact-an updated review.Environ Sci Pollut Res Int. 2021 May;28(18):22241-22264. doi: 10.1007/s11356-021-13018-1. Epub 2021 Mar 18. Environ Sci Pollut Res Int. 2021. PMID: 33733422 Free PMC article. Review.
References
-
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

