Genome-wide identification of miRNAs and their targets during early somatic embryogenesis in Dimocarpus longan Lour
- PMID: 32170163
- PMCID: PMC7069941
- DOI: 10.1038/s41598-020-60946-y
Genome-wide identification of miRNAs and their targets during early somatic embryogenesis in Dimocarpus longan Lour
Abstract
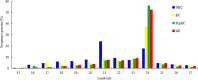
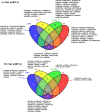
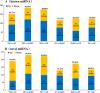
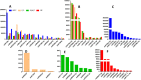
miRNAs are endogenous regulatory factors that play pivotal roles in post-transcriptional regulation. However, their specific roles in early somatic embryogenesis (SE) remain unclear. Study of the SE system is fundamental for clarifying the molecular mechanisms in Dimocarpus longan. We identified 289 known miRNAs from 106 different miRNA families and 1087 novel miRNAs during early longan SE, including embryogenic callus (EC), incomplete pro-embryogenic culture (ICpEC), globular embryo (GE), and non-embryogenic callus (NEC). The abundances of known miRNAs were concentrated in GE. The differentially expression (DE) miRNAs showed five expression patterns during early SE. Largely miRNAs were expressed highly and specially in EC, ICpEC, and GE, respectively. Some miRNAs and putative target genes were enriched in lignin metabolism. Most potential targets were related to the pathways of plant hormone signal transduction, alternative splicing, tyrosine metabolism and sulfur metabolism in early longan SE. The regulatory relationships between dlo-miR166a-3p and DlHD-zip8, dlo-miR397a and DlLAC7, dlo-miR408-3p and DlLAC12 were confirmed by RNA ligase-mediated rapid amplification of cDNA ends. The expression patterns of eight DE miRNAs detected by qRT-PCR were consistent with RNA-seq. Finally, the miRNA regulatory network in early SE was constructed, which provided new insight into molecular mechanism of early SE in longan.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Comparative analysis reveals dynamic changes in miRNAs and their targets and expression during somatic embryogenesis in longan (Dimocarpus longan Lour.).PLoS One. 2013 Apr 11;8(4):e60337. doi: 10.1371/journal.pone.0060337. Print 2013. PLoS One. 2013. PMID: 23593197 Free PMC article.
-
Global scale transcriptome analysis reveals differentially expressed genes involve in early somatic embryogenesis in Dimocarpus longan Lour.BMC Genomics. 2020 Jan 2;21(1):4. doi: 10.1186/s12864-019-6393-7. BMC Genomics. 2020. PMID: 31898486 Free PMC article.
-
Genome-wide identification of miRNAs and targets associated with cell wall biosynthesis: Differential roles of dlo-miR397a and dlo-miR408-3p during early somatic embryogenesis in longan.Plant Sci. 2022 Oct;323:111372. doi: 10.1016/j.plantsci.2022.111372. Epub 2022 Jul 18. Plant Sci. 2022. PMID: 35863557
-
The role of miRNA in somatic embryogenesis.Genomics. 2019 Sep;111(5):1026-1033. doi: 10.1016/j.ygeno.2018.11.022. Epub 2018 Nov 23. Genomics. 2019. PMID: 30476555 Review.
-
Research Tools for the Functional Genomics of Plant miRNAs During Zygotic and Somatic Embryogenesis.Int J Mol Sci. 2020 Jul 14;21(14):4969. doi: 10.3390/ijms21144969. Int J Mol Sci. 2020. PMID: 32674459 Free PMC article. Review.
Cited by
-
Physiological and Molecular Responses of Pyrus pyraster Seedlings to Salt Treatment Analyzed by miRNA and Cytochrome P450 Gene-Based Markers.Plants (Basel). 2024 Jan 16;13(2):261. doi: 10.3390/plants13020261. Plants (Basel). 2024. PMID: 38256814 Free PMC article.
-
Small Non-Coding RNAs at the Crossroads of Regulatory Pathways Controlling Somatic Embryogenesis in Seed Plants.Plants (Basel). 2021 Mar 9;10(3):504. doi: 10.3390/plants10030504. Plants (Basel). 2021. PMID: 33803088 Free PMC article. Review.
-
MicroRNA Zma-miR528 Versatile Regulation on Target mRNAs during Maize Somatic Embryogenesis.Int J Mol Sci. 2021 May 18;22(10):5310. doi: 10.3390/ijms22105310. Int J Mol Sci. 2021. PMID: 34069987 Free PMC article.
-
Identification of Novel miRNAs and Their Target Genes in the Response to Abscisic Acid in Arabidopsis.Int J Mol Sci. 2021 Jul 1;22(13):7153. doi: 10.3390/ijms22137153. Int J Mol Sci. 2021. PMID: 34281207 Free PMC article.
-
Hypermethylation of Auxin-Responsive Motifs in the Promoters of the Transcription Factor Genes Accompanies the Somatic Embryogenesis Induction in Arabidopsis.Int J Mol Sci. 2020 Sep 18;21(18):6849. doi: 10.3390/ijms21186849. Int J Mol Sci. 2020. PMID: 32961931 Free PMC article.
References
-
- Yang X, et al. Regulation of somatic embryogenesis in higher plants. Crit. Revi Plant. Sciences. 2010;29:36–57. doi: 10.1080/07352680903436291. - DOI
-
- Chen, X. M. MicroRNA metabolism in plants[M]//Rna Interference. Springer, Berlin, Heidelberg, :117–136 (2008).
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

