Phosphorylated STAT3 (Tyr705) as a biomarker of response to pimozide treatment in triple-negative breast cancer
- PMID: 32164483
- PMCID: PMC7515519
- DOI: 10.1080/15384047.2020.1726718
Phosphorylated STAT3 (Tyr705) as a biomarker of response to pimozide treatment in triple-negative breast cancer
Abstract
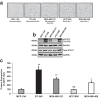
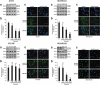
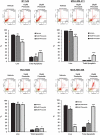
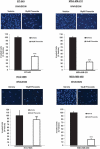
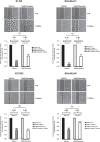
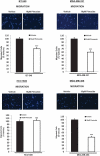
Triple-negative breast cancer (TNBC) displays an aggressive clinical course, heightened metastatic potential, and is linked to poor survival rates. Through its lack of expression of the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2), this subtype remains unresponsive to traditional targeted therapies. Undesirable and sometimes life-threatening side effects associated with current chemotherapeutic agents warrant the development of more targeted treatment options. Targeting signal transducer and activator of transcription 3 (STAT3), a transcription factor implicated in breast cancer (BCa) progression, has proven to be an efficient approach to halt cancer growth in vitro and in vivo. Currently, there are no FDA-approved STAT3 inhibitors for TNBC. Although pimozide, a FDA-approved antipsychotic drug, has been attributed a role as a STAT3 inhibitor in several cancers, its role on this pathway remains unexplored in TNBC. As a "one size fits all" approach cannot be applied to TNBC therapies due to the heterogeneous nature of this aggressive cancer, we hypothesized that STAT3 could be a novel biomarker of response to guide pimozide therapy. Using human cell lines representative of four TNBC subtypes (basal-like 1, basal-like 2, mesenchymal-like, mesenchymal stem-like), our current report demonstrates that pimozide significantly reduced their invasion and migration, an effect that was predicted by STAT3 phosphorylation on tyrosine residue 705 (Tyr705). Mechanistically, phosphorylated STAT3 (Tyr705) inhibition resulting from pimozide treatment caused a downregulation of downstream transcriptional targets such as matrix metalloproteinase-9 (MMP-9) and vimentin, both implicated in invasion and migration. The identification of biomarkers of response to TNBC treatments is an active area of research in the field of precision medicine and our results propose phosphorylated STAT3 (Tyr705) as a novel biomarker to guide pimozide treatment as an inhibitor of invasion and migration.
Keywords: STAT3; biomarker; drug repurposing; pimozide; precision Medicine; triple-negative breast cancer.
Figures


Similar articles
-
STAT3 as a potential therapeutic target in triple negative breast cancer: a systematic review.J Exp Clin Cancer Res. 2019 May 14;38(1):195. doi: 10.1186/s13046-019-1206-z. J Exp Clin Cancer Res. 2019. PMID: 31088482 Free PMC article.
-
Genomic regulation of invasion by STAT3 in triple negative breast cancer.Oncotarget. 2017 Jan 31;8(5):8226-8238. doi: 10.18632/oncotarget.14153. Oncotarget. 2017. PMID: 28030809 Free PMC article.
-
PDSS1-Mediated Activation of CAMK2A-STAT3 Signaling Promotes Metastasis in Triple-Negative Breast Cancer.Cancer Res. 2021 Nov 1;81(21):5491-5505. doi: 10.1158/0008-5472.CAN-21-0747. Epub 2021 Aug 18. Cancer Res. 2021. PMID: 34408002
-
Stress-induced EGF receptor signaling through STAT3 and tumor progression in triple-negative breast cancer.Mol Cell Endocrinol. 2017 Aug 15;451:24-30. doi: 10.1016/j.mce.2017.01.013. Epub 2017 Jan 12. Mol Cell Endocrinol. 2017. PMID: 28088463 Free PMC article. Review.
-
Targeting Different Pathways Using Novel Combination Therapy in Triple Negative Breast Cancer.Curr Cancer Drug Targets. 2020;20(8):586-602. doi: 10.2174/1570163817666200518081955. Curr Cancer Drug Targets. 2020. PMID: 32418525 Review.
Cited by
-
Combination of CTLA-4 blockade with MUC1 mRNA nanovaccine induces enhanced anti-tumor CTL activity by modulating tumor microenvironment of triple negative breast cancer.Transl Oncol. 2022 Jan;15(1):101298. doi: 10.1016/j.tranon.2021.101298. Epub 2021 Dec 4. Transl Oncol. 2022. PMID: 34875483 Free PMC article.
-
STAT5a Confers Doxorubicin Resistance to Breast Cancer by Regulating ABCB1.Front Oncol. 2021 Jul 15;11:697950. doi: 10.3389/fonc.2021.697950. eCollection 2021. Front Oncol. 2021. PMID: 34336684 Free PMC article.
-
Drug Repurposing to Enhance Antitumor Response to PD-1/PD-L1 Immune Checkpoint Inhibitors.Cancers (Basel). 2022 Jul 11;14(14):3368. doi: 10.3390/cancers14143368. Cancers (Basel). 2022. PMID: 35884428 Free PMC article. Review.
-
Network medicine based approach for identifying the type 2 diabetes, osteoarthritis and triple negative breast cancer interactome: Finding the hub of hub genes.Heliyon. 2024 Aug 22;10(17):e36650. doi: 10.1016/j.heliyon.2024.e36650. eCollection 2024 Sep 15. Heliyon. 2024. PMID: 39281650 Free PMC article.
-
Pimozide-loaded nanostructured lipid carriers: Repurposing strategy against lung cancer.Sci Prog. 2024 Oct-Dec;107(4):368504241296304. doi: 10.1177/00368504241296304. Sci Prog. 2024. PMID: 39497512 Free PMC article.
References
-
- Uscanga-Perales GI, Santuario-Facio SK, Ortiz-Lopez R. Triple negative breast cancer: deciphering the biology and heterogeneity. Med Universitaria. 2016;18:105–114. doi:10.1016/j.rmu.2016.05.007. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
