Single cell transcriptomics identifies focal segmental glomerulosclerosis remission endothelial biomarker
- PMID: 32107344
- PMCID: PMC7213795
- DOI: 10.1172/jci.insight.133267
Single cell transcriptomics identifies focal segmental glomerulosclerosis remission endothelial biomarker
Abstract
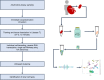
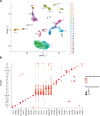
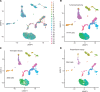
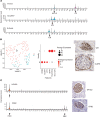
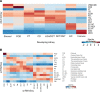
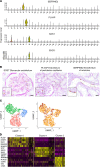
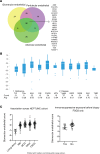
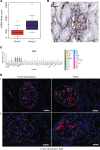
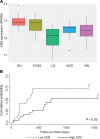
To define cellular mechanisms underlying kidney function and failure, the KPMP analyzes biopsy tissue in a multicenter research network to build cell-level process maps of the kidney. This study aimed to establish a single cell RNA sequencing strategy to use cell-level transcriptional profiles from kidney biopsies in KPMP to define molecular subtypes in glomerular diseases. Using multiple sources of adult human kidney reference tissue samples, 22,268 single cell profiles passed KPMP quality control parameters. Unbiased clustering resulted in 31 distinct cell clusters that were linked to kidney and immune cell types using specific cell markers. Focusing on endothelial cell phenotypes, in silico and in situ hybridization methods assigned 3 discrete endothelial cell clusters to distinct renal vascular beds. Transcripts defining glomerular endothelial cells (GEC) were evaluated in biopsies from patients with 10 different glomerular diseases in the NEPTUNE and European Renal cDNA Bank (ERCB) cohort studies. Highest GEC scores were observed in patients with focal segmental glomerulosclerosis (FSGS). Molecular endothelial signatures suggested 2 distinct FSGS patient subgroups with α-2 macroglobulin (A2M) as a key downstream mediator of the endothelial cell phenotype. Finally, glomerular A2M transcript levels associated with lower proteinuria remission rates, linking endothelial function with long-term outcome in FSGS.
Keywords: Bioinformatics; Cell Biology; Chronic kidney disease; Nephrology; endothelial cells.
Conflict of interest statement
Figures


Similar articles
-
Glomerular endothelial cell heterogeneity in Alport syndrome.Sci Rep. 2020 Jul 10;10(1):11414. doi: 10.1038/s41598-020-67588-0. Sci Rep. 2020. PMID: 32651395 Free PMC article.
-
Glomerular endothelial cell injury and focal segmental glomerulosclerosis lesion in idiopathic membranous nephropathy.PLoS One. 2015 Apr 15;10(4):e0116700. doi: 10.1371/journal.pone.0116700. eCollection 2015. PLoS One. 2015. PMID: 25875837 Free PMC article.
-
Cellular Senescence Is Associated with Faster Progression of Focal Segmental Glomerulosclerosis.Am J Nephrol. 2020;51(12):950-958. doi: 10.1159/000511560. Epub 2021 Jan 13. Am J Nephrol. 2020. PMID: 33440379
-
Update on Recurrent Focal Segmental Glomerulosclerosis in Kidney Transplantation.Nephron. 2020;144 Suppl 1:65-70. doi: 10.1159/000510748. Epub 2020 Dec 1. Nephron. 2020. PMID: 33260184 Review.
-
Secondary Focal Segmental Glomerulosclerosis: From Podocyte Injury to Glomerulosclerosis.Biomed Res Int. 2016;2016:1630365. doi: 10.1155/2016/1630365. Epub 2016 Mar 21. Biomed Res Int. 2016. PMID: 27088082 Free PMC article. Review.
Cited by
-
Identification of kidney cell types in scRNA-seq and snRNA-seq data using machine learning algorithms.Heliyon. 2024 Sep 27;10(19):e38567. doi: 10.1016/j.heliyon.2024.e38567. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39403515 Free PMC article.
-
Epithelial Na + Channels Function as Extracellular Sensors.Compr Physiol. 2024 Mar 29;14(2):1-41. doi: 10.1002/cphy.c230015. Compr Physiol. 2024. PMID: 39109974 Review.
-
Varying Selection Pressure for a Na+ Sensing Site in Epithelial Na+ Channel Subunits Reflect Divergent Roles in Na+ Homeostasis.Mol Biol Evol. 2024 Aug 2;41(8):msae162. doi: 10.1093/molbev/msae162. Mol Biol Evol. 2024. PMID: 39101592 Free PMC article.
-
Single-cell RNA sequencing in diabetic kidney disease: a literature review.Ren Fail. 2024 Dec;46(2):2387428. doi: 10.1080/0886022X.2024.2387428. Epub 2024 Aug 4. Ren Fail. 2024. PMID: 39099183 Free PMC article. Review.
-
Integrated multi-omics with machine learning to uncover the intricacies of kidney disease.Brief Bioinform. 2024 Jul 25;25(5):bbae364. doi: 10.1093/bib/bbae364. Brief Bioinform. 2024. PMID: 39082652 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

