TRPM3_miR-204: a complex locus for eye development and disease
- PMID: 32070426
- PMCID: PMC7027284
- DOI: 10.1186/s40246-020-00258-4
TRPM3_miR-204: a complex locus for eye development and disease
Abstract
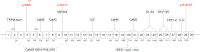
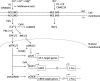
First discovered in a light-sensitive retinal mutant of Drosophila, the transient receptor potential (TRP) superfamily of non-selective cation channels serve as polymodal cellular sensors that participate in diverse physiological processes across the animal kingdom including the perception of light, temperature, pressure, and pain. TRPM3 belongs to the melastatin sub-family of TRP channels and has been shown to function as a spontaneous calcium channel, with permeability to other cations influenced by alternative splicing and/or non-canonical channel activity. Activators of TRPM3 channels include the neurosteroid pregnenolone sulfate, calmodulin, phosphoinositides, and heat, whereas inhibitors include certain drugs, plant-derived metabolites, and G-protein subunits. Activation of TRPM3 channels at the cell membrane elicits a signal transduction cascade of mitogen-activated kinases and stimulus response transcription factors. The mammalian TRPM3 gene hosts a non-coding microRNA gene specifying miR-204 that serves as both a tumor suppressor and a negative regulator of post-transcriptional gene expression during eye development in vertebrates. Ocular co-expression of TRPM3 and miR-204 is upregulated by the paired box 6 transcription factor (PAX6) and mutations in all three corresponding genes underlie inherited forms of eye disease in humans including early-onset cataract, retinal dystrophy, and coloboma. This review outlines the genomic and functional complexity of the TRPM3_miR-204 locus in mammalian eye development and disease.
Keywords: Eye development; Eye disease; MicroRNA; TRP channel.
Conflict of interest statement
The author declares that he has no competing interests.
Figures



Similar articles
-
Transient receptor potential TRPM3 channels: Pharmacology, signaling, and biological functions.Pharmacol Res. 2017 Oct;124:92-99. doi: 10.1016/j.phrs.2017.07.014. Epub 2017 Jul 16. Pharmacol Res. 2017. PMID: 28720517 Review.
-
Transient receptor potential melastatin-3 (TRPM3)-induced activation of AP-1 requires Ca2+ ions and the transcription factors c-Jun, ATF2, and ternary complex factor.Mol Pharmacol. 2015 Apr;87(4):617-28. doi: 10.1124/mol.114.095695. Epub 2015 Jan 9. Mol Pharmacol. 2015. PMID: 25576487
-
Disease-associated mutations in the human TRPM3 render the channel overactive via two distinct mechanisms.Elife. 2020 Apr 28;9:e55634. doi: 10.7554/eLife.55634. Elife. 2020. PMID: 32343227 Free PMC article.
-
TRPM3.Handb Exp Pharmacol. 2014;222:427-59. doi: 10.1007/978-3-642-54215-2_17. Handb Exp Pharmacol. 2014. PMID: 24756716 Review.
-
Activation and inhibition of transient receptor potential TRPM3-induced gene transcription.Br J Pharmacol. 2014 May;171(10):2645-58. doi: 10.1111/bph.12524. Br J Pharmacol. 2014. PMID: 24895737 Free PMC article.
Cited by
-
Functions of TRPs in retinal tissue in physiological and pathological conditions.Front Mol Neurosci. 2024 Sep 25;17:1459083. doi: 10.3389/fnmol.2024.1459083. eCollection 2024. Front Mol Neurosci. 2024. PMID: 39386050 Free PMC article. Review.
-
miR-204-containing exosomes ameliorate GVHD-associated dry eye disease.Sci Adv. 2022 Jan 14;8(2):eabj9617. doi: 10.1126/sciadv.abj9617. Epub 2022 Jan 12. Sci Adv. 2022. PMID: 35020440 Free PMC article.
-
miR-204: Molecular Regulation and Role in Cardiovascular and Renal Diseases.Hypertension. 2021 Aug;78(2):270-281. doi: 10.1161/HYPERTENSIONAHA.121.14536. Epub 2021 Jun 28. Hypertension. 2021. PMID: 34176282 Free PMC article. Review.
-
Unraveling the Roles of miR-204-5p and HMGA2 in Papillary Thyroid Cancer Tumorigenesis.Int J Mol Sci. 2023 Jun 28;24(13):10764. doi: 10.3390/ijms241310764. Int J Mol Sci. 2023. PMID: 37445942 Free PMC article.
-
The role of TRPV4 channels in ocular function and pathologies.Exp Eye Res. 2020 Dec;201:108257. doi: 10.1016/j.exer.2020.108257. Epub 2020 Sep 29. Exp Eye Res. 2020. PMID: 32979394 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

