Structural dynamics of the human COP9 signalosome revealed by cross-linking mass spectrometry and integrative modeling
- PMID: 32034103
- PMCID: PMC7049115
- DOI: 10.1073/pnas.1915542117
Structural dynamics of the human COP9 signalosome revealed by cross-linking mass spectrometry and integrative modeling
Abstract
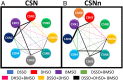
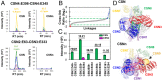
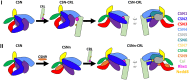
The COP9 signalosome (CSN) is an evolutionarily conserved eight-subunit (CSN1-8) protein complex that controls protein ubiquitination by deneddylating Cullin-RING E3 ligases (CRLs). The activation and function of CSN hinges on its structural dynamics, which has been challenging to decipher by conventional tools. Here, we have developed a multichemistry cross-linking mass spectrometry approach enabled by three mass spectometry-cleavable cross-linkers to generate highly reliable cross-link data. We applied this approach with integrative structure modeling to determine the interaction and structural dynamics of CSN with the recently discovered ninth subunit, CSN9, in solution. Our results determined the localization of CSN9 binding sites and revealed CSN9-dependent structural changes of CSN. Together with biochemical analysis, we propose a structural model in which CSN9 binding triggers CSN to adopt a configuration that facilitates CSN-CRL interactions, thereby augmenting CSN deneddylase activity. Our integrative structure analysis workflow can be generalized to define in-solution architectures of dynamic protein complexes that remain inaccessible to other approaches.
Keywords: COP9 signalosome; architectures of protein complexes; cross-linking mass spectrometry; integrative structure modeling; structural dynamics.
Conflict of interest statement
The authors declare no competing interest.
Figures


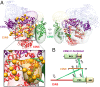
Similar articles
-
Are Inositol Polyphosphates the Missing Link in Dynamic Cullin RING Ligase Regulation by the COP9 Signalosome?Biomolecules. 2019 Aug 7;9(8):349. doi: 10.3390/biom9080349. Biomolecules. 2019. PMID: 31394817 Free PMC article. Review.
-
Cullin-RING Ligase Regulation by the COP9 Signalosome: Structural Mechanisms and New Physiologic Players.Adv Exp Med Biol. 2020;1217:47-60. doi: 10.1007/978-981-15-1025-0_4. Adv Exp Med Biol. 2020. PMID: 31898221 Review.
-
Basis for metabolite-dependent Cullin-RING ligase deneddylation by the COP9 signalosome.Proc Natl Acad Sci U S A. 2020 Feb 25;117(8):4117-4124. doi: 10.1073/pnas.1911998117. Epub 2020 Feb 11. Proc Natl Acad Sci U S A. 2020. PMID: 32047038 Free PMC article.
-
IKK-Mediated Regulation of the COP9 Signalosome via Phosphorylation of CSN5.J Proteome Res. 2020 Mar 6;19(3):1119-1130. doi: 10.1021/acs.jproteome.9b00626. Epub 2020 Feb 3. J Proteome Res. 2020. PMID: 31950832 Free PMC article.
-
Role of the COP9 Signalosome (CSN) in Cardiovascular Diseases.Biomolecules. 2019 Jun 5;9(6):217. doi: 10.3390/biom9060217. Biomolecules. 2019. PMID: 31195722 Free PMC article. Review.
Cited by
-
Spatially resolved profiling of protein conformation and interactions by biocompatible chemical cross-linking in living cells.Nat Commun. 2024 Sep 27;15(1):8331. doi: 10.1038/s41467-024-52558-1. Nat Commun. 2024. PMID: 39333085 Free PMC article.
-
Crosslinking mass spectrometry: A link between structural biology and systems biology.Protein Sci. 2021 Apr;30(4):773-784. doi: 10.1002/pro.4045. Epub 2021 Mar 6. Protein Sci. 2021. PMID: 33594738 Free PMC article. Review.
-
Integrative structure determination of histones H3 and H4 using genetic interactions.FEBS J. 2023 May;290(10):2565-2575. doi: 10.1111/febs.16435. Epub 2022 Apr 3. FEBS J. 2023. PMID: 35298864 Free PMC article.
-
The protein organization of a red blood cell.Cell Rep. 2022 Jul 19;40(3):111103. doi: 10.1016/j.celrep.2022.111103. Cell Rep. 2022. PMID: 35858567 Free PMC article.
-
A Framework for Stochastic Optimization of Parameters for Integrative Modeling of Macromolecular Assemblies.Life (Basel). 2021 Nov 5;11(11):1183. doi: 10.3390/life11111183. Life (Basel). 2021. PMID: 34833059 Free PMC article.
References
-
- Wei N., Deng X. W., The COP9 signalosome. Annu. Rev. Cell Dev. Biol. 19, 261–286 (2003). - PubMed
-
- Wolf D. A., Zhou C., Wee S., The COP9 signalosome: An assembly and maintenance platform for cullin ubiquitin ligases? Nat. Cell Biol. 5, 1029–1033 (2003). - PubMed
-
- Wei N., Serino G., Deng X. W., The COP9 signalosome: More than a protease. Trends Biochem. Sci. 33, 592–600 (2008). - PubMed
-
- Cope G. A., et al. , Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298, 608–611 (2002). - PubMed
-
- Deshaies R. J., Joazeiro C. A., RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

