Actin reduction by MsrB2 is a key component of the cytokinetic abscission checkpoint and prevents tetraploidy
- PMID: 32029597
- PMCID: PMC7049109
- DOI: 10.1073/pnas.1911629117
Actin reduction by MsrB2 is a key component of the cytokinetic abscission checkpoint and prevents tetraploidy
Abstract
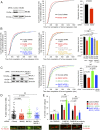
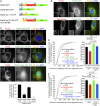
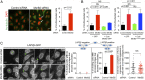
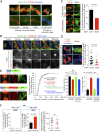
Abscission is the terminal step of cytokinesis leading to the physical separation of the daughter cells. In response to the abnormal presence of lagging chromatin between dividing cells, an evolutionarily conserved abscission/NoCut checkpoint delays abscission and prevents formation of binucleated cells by stabilizing the cytokinetic intercellular bridge (ICB). How this bridge is stably maintained for hours while the checkpoint is activated is poorly understood and has been proposed to rely on F-actin in the bridge region. Here, we show that actin polymerization is indeed essential for stabilizing the ICB when lagging chromatin is present, but not in normal dividing cells. Mechanistically, we found that a cytosolic pool of human methionine sulfoxide reductase B2 (MsrB2) is strongly recruited at the midbody in response to the presence of lagging chromatin and functions within the ICB to promote actin polymerization there. Consistently, in MsrB2-depleted cells, F-actin levels are decreased in ICBs, and dividing cells with lagging chromatin become binucleated as a consequence of unstable bridges. We further demonstrate that MsrB2 selectively reduces oxidized actin monomers and thereby counteracts MICAL1, an enzyme known to depolymerize actin filaments by direct oxidation. Finally, MsrB2 colocalizes and genetically interacts with the checkpoint components Aurora B and ANCHR, and the abscission delay upon checkpoint activation by nuclear pore defects also depends on MsrB2. Altogether, this work reveals that actin reduction by MsrB2 is a key component of the abscission checkpoint that favors F-actin polymerization and limits tetraploidy, a starting point for tumorigenesis.
Keywords: abscission checkpoint; actin; cytokinesis; cytoskeleton; oxidoreduction.
Conflict of interest statement
The authors declare no competing interest.
Figures

Similar articles
-
DENND2B activates Rab35 at the intercellular bridge, regulating cytokinetic abscission and tetraploidy.Cell Rep. 2023 Jul 25;42(7):112795. doi: 10.1016/j.celrep.2023.112795. Epub 2023 Jul 16. Cell Rep. 2023. PMID: 37454296
-
Oxidation of F-actin controls the terminal steps of cytokinesis.Nat Commun. 2017 Feb 23;8:14528. doi: 10.1038/ncomms14528. Nat Commun. 2017. PMID: 28230050 Free PMC article.
-
The Abscission Checkpoint: A Guardian of Chromosomal Stability.Cells. 2021 Nov 29;10(12):3350. doi: 10.3390/cells10123350. Cells. 2021. PMID: 34943860 Free PMC article. Review.
-
Capping protein regulates actin dynamics during cytokinetic midbody maturation.Proc Natl Acad Sci U S A. 2018 Feb 27;115(9):2138-2143. doi: 10.1073/pnas.1722281115. Epub 2018 Feb 8. Proc Natl Acad Sci U S A. 2018. PMID: 29439200 Free PMC article.
-
The Abscission Checkpoint: Making It to the Final Cut.Trends Cell Biol. 2017 Jan;27(1):1-11. doi: 10.1016/j.tcb.2016.10.001. Epub 2016 Oct 31. Trends Cell Biol. 2017. PMID: 27810282 Review.
Cited by
-
Minor Kinases with Major Roles in Cytokinesis Regulation.Cells. 2022 Nov 17;11(22):3639. doi: 10.3390/cells11223639. Cells. 2022. PMID: 36429067 Free PMC article. Review.
-
Mechanics and regulation of cytokinetic abscission.Front Cell Dev Biol. 2022 Nov 24;10:1046617. doi: 10.3389/fcell.2022.1046617. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36506096 Free PMC article. Review.
-
The Last Chance Saloon.Front Cell Dev Biol. 2021 May 14;9:671297. doi: 10.3389/fcell.2021.671297. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34055803 Free PMC article. Review.
-
Actin filament oxidation by MICAL1 suppresses protections from cofilin-induced disassembly.EMBO Rep. 2021 Feb 3;22(2):e50965. doi: 10.15252/embr.202050965. Epub 2021 Jan 4. EMBO Rep. 2021. PMID: 33393173 Free PMC article.
-
Preserving Genome Integrity: Unveiling the Roles of ESCRT Machinery.Cells. 2024 Aug 5;13(15):1307. doi: 10.3390/cells13151307. Cells. 2024. PMID: 39120335 Free PMC article. Review.
References
-
- Green R. A., Paluch E., Oegema K., Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 28, 29–58 (2012). - PubMed
-
- Mierzwa B., Gerlich D. W., Cytokinetic abscission: Molecular mechanisms and temporal control. Dev. Cell 31, 525–538 (2014). - PubMed
-
- Frémont S., Echard A., Membrane traffic in the late steps of cytokinesis. Curr. Biol. 28, R458–R470 (2018). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

