Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV
- PMID: 32022370
- PMCID: PMC7162020
- DOI: 10.1002/cbic.202000047
Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV
Abstract
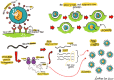
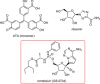
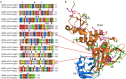
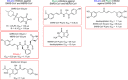
With the current trajectory of the 2019-nCoV outbreak unknown, public health and medicinal measures will both be needed to contain spreading of the virus and to optimize patient outcomes. Although little is known about the virus, an examination of the genome sequence shows strong homology with its better-studied cousin, SARS-CoV. The spike protein used for host cell infection shows key nonsynonymous mutations that might hamper the efficacy of previously developed therapeutics but remains a viable target for the development of biologics and macrocyclic peptides. Other key drug targets, including RNA-dependent RNA polymerase and coronavirus main proteinase (3CLpro), share a strikingly high (>95 %) homology to SARS-CoV. Herein, we suggest four potential drug candidates (an ACE2-based peptide, remdesivir, 3CLpro-1 and a novel vinylsulfone protease inhibitor) that could be used to treat patients suffering with the 2019-nCoV. We also summarize previous efforts into drugging these targets and hope to help in the development of broad-spectrum anti-coronaviral agents for future epidemics.
Keywords: 2019-nCoV; 3CLpro; RdRp; SARS; antiviral agents; coronavirus; spike proteins.
© 2020 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
Figures



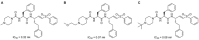
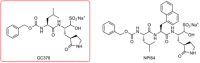
Update of
-
Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV.ChemRxiv [Preprint]. 2020 Jan 27. doi: 10.26434/chemrxiv.11728983. ChemRxiv. 2020. Update in: Chembiochem. 2020 Mar 2;21(5):730-738. doi: 10.1002/cbic.202000047. PMID: 32511285 Free PMC article. Updated. Preprint.
Similar articles
-
Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV.ChemRxiv [Preprint]. 2020 Jan 27. doi: 10.26434/chemrxiv.11728983. ChemRxiv. 2020. Update in: Chembiochem. 2020 Mar 2;21(5):730-738. doi: 10.1002/cbic.202000047. PMID: 32511285 Free PMC article. Updated. Preprint.
-
Targeting the Dimerization of the Main Protease of Coronaviruses: A Potential Broad-Spectrum Therapeutic Strategy.ACS Comb Sci. 2020 Jun 8;22(6):297-305. doi: 10.1021/acscombsci.0c00058. Epub 2020 May 27. ACS Comb Sci. 2020. PMID: 32402186 Review.
-
Deep Learning Based Drug Screening for Novel Coronavirus 2019-nCov.Interdiscip Sci. 2020 Sep;12(3):368-376. doi: 10.1007/s12539-020-00376-6. Epub 2020 Jun 1. Interdiscip Sci. 2020. PMID: 32488835 Free PMC article.
-
Novel Drugs Targeting the SARS-CoV-2/COVID-19 Machinery.Curr Top Med Chem. 2020;20(16):1423-1433. doi: 10.2174/1568026620999200517043137. Curr Top Med Chem. 2020. PMID: 32416679
-
Design and Evaluation of Anti-SARS-Coronavirus Agents Based on Molecular Interactions with the Viral Protease.Molecules. 2020 Aug 27;25(17):3920. doi: 10.3390/molecules25173920. Molecules. 2020. PMID: 32867349 Free PMC article. Review.
Cited by
-
Peptide-Based Inhibitors of Protein-Protein Interactions (PPIs): A Case Study on the Interaction Between SARS-CoV-2 Spike Protein and Human Angiotensin-Converting Enzyme 2 (hACE2).Biomedicines. 2024 Oct 16;12(10):2361. doi: 10.3390/biomedicines12102361. Biomedicines. 2024. PMID: 39457672 Free PMC article. Review.
-
Assay Development and Validation for Innovative Antiviral Development Targeting the N-Terminal Autoprocessing of SARS-CoV-2 Main Protease Precursors.Viruses. 2024 Jul 29;16(8):1218. doi: 10.3390/v16081218. Viruses. 2024. PMID: 39205192 Free PMC article.
-
Phytoconstituents of Citrus limon (Lemon) as Potential Inhibitors Against Multi Targets of SARS-CoV-2 by Use of Molecular Modelling and In Vitro Determination Approaches.ChemistryOpen. 2024 Oct;13(10):e202300198. doi: 10.1002/open.202300198. Epub 2024 Jun 21. ChemistryOpen. 2024. PMID: 39031747 Free PMC article.
-
SARS-CoV-2 Main Protease Inhibitors That Leverage Unique Interactions with the Solvent Exposed S3 Site of the Enzyme.ACS Med Chem Lett. 2024 May 20;15(6):950-957. doi: 10.1021/acsmedchemlett.4c00146. eCollection 2024 Jun 13. ACS Med Chem Lett. 2024. PMID: 38894905 Free PMC article.
-
A Narrative Overview of Coronavirus Infection: Clinical Signs and Symptoms, Viral Entry and Replication, Treatment Modalities, and Management.Curr Top Med Chem. 2024;24(21):1883-1916. doi: 10.2174/0115680266296095240529114058. Curr Top Med Chem. 2024. PMID: 38859776 Review.
References
-
- Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., Si H. R., Zhu Y., Li B., Huang C. L., Chen H. D., Chen J., Luo Y., Guo H., Jiang R. D., Liu M. Q., Chen Y., Shen X. R., Wang X., Zheng X. S., Zhao K., Chen Q. J., Deng F., Liu L. L., Yan B., Zhan F. X., Wang Y. Y., Xiao G., Shi Z., bioRxiv 2020, https://www.biorxiv.org/content/10.1101/2020.01.22.914952v2 (accessed Jan. 25, 2020). - DOI
-
- Li F., Li W., Farzan M., Harrison S. C., Science 2005, 309, 1864–1868. - PubMed
-
- Dong N., Yang X., Ye L., Chen K., Chan E. W. C., Yang M., Chen S., bioRxiv 2020, https://www.biorxiv.org/content/10.1101/2020.01.20.913368v2 (accessed Jan. 25, 2020). - DOI
-
- Ziebuhr J., Snijder E. J., Gorbalenya A. E., J. Gen. Virol. 2000, 81, 853–879. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

