RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak
- PMID: 32020836
- PMCID: PMC7033720
- DOI: 10.1080/22221751.2020.1725399
RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak
Abstract
From December 2019, an outbreak of unusual pneumonia was reported in Wuhan with many cases linked to Huanan Seafood Market that sells seafood as well as live exotic animals. We investigated two patients who developed acute respiratory syndromes after independent contact history with this market. The two patients shared common clinical features including fever, cough, and multiple ground-glass opacities in the bilateral lung field with patchy infiltration. Here, we highlight the use of a low-input metagenomic next-generation sequencing (mNGS) approach on RNA extracted from bronchoalveolar lavage fluid (BALF). It rapidly identified a novel coronavirus (named 2019-nCoV according to World Health Organization announcement) which was the sole pathogens in the sample with very high abundance level (1.5% and 0.62% of total RNA sequenced). The entire viral genome is 29,881 nt in length (GenBank MN988668 and MN988669, Sequence Read Archive database Bioproject accession PRJNA601736) and is classified into β-coronavirus genus. Phylogenetic analysis indicates that 2019-nCoV is close to coronaviruses (CoVs) circulating in Rhinolophus (Horseshoe bats), such as 98.7% nucleotide identity to partial RdRp gene of bat coronavirus strain BtCoV/4991 (GenBank KP876546, 370 nt sequence of RdRp and lack of other genome sequence) and 87.9% nucleotide identity to bat coronavirus strain bat-SL-CoVZC45 and bat-SL-CoVZXC21. Evolutionary analysis based on ORF1a/1b, S, and N genes also suggests 2019-nCoV is more likely a novel CoV independently introduced from animals to humans.
Keywords: 2019-nCoV; Wuhan pneumonia; metagenomic next-generation sequencing; phylogenetic analyses; virus evolution.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

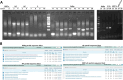
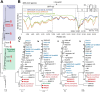
Comment in
-
Antihypertensive treatment with ACEI/ARB of patients with COVID-19 complicated by hypertension.Hypertens Res. 2020 Jun;43(6):588-590. doi: 10.1038/s41440-020-0433-1. Epub 2020 Mar 30. Hypertens Res. 2020. PMID: 32231220 Free PMC article. No abstract available.
Similar articles
-
Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding.Lancet. 2020 Feb 22;395(10224):565-574. doi: 10.1016/S0140-6736(20)30251-8. Epub 2020 Jan 30. Lancet. 2020. PMID: 32007145 Free PMC article.
-
Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan.Emerg Microbes Infect. 2020 Jan 28;9(1):221-236. doi: 10.1080/22221751.2020.1719902. eCollection 2020. Emerg Microbes Infect. 2020. PMID: 31987001 Free PMC article.
-
[Etiology of epidemic outbreaks COVID-19 on Wuhan, Hubei province, Chinese People Republic associated with 2019-nCoV (Nidovirales, Coronaviridae, Coronavirinae, Betacoronavirus, Subgenus Sarbecovirus): lessons of SARS-CoV outbreak.].Vopr Virusol. 2020;65(1):6-15. doi: 10.36233/0507-4088-2020-65-1-6-15. Vopr Virusol. 2020. PMID: 32496715 Review. Russian.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
-
Cross-species transmission of the newly identified coronavirus 2019-nCoV.J Med Virol. 2020 Apr;92(4):433-440. doi: 10.1002/jmv.25682. J Med Virol. 2020. PMID: 31967321 Free PMC article.
Cited by
-
Drug Repurposing of Itraconazole and Estradiol Benzoate against COVID-19 by Blocking SARS-CoV-2 Spike Protein-Mediated Membrane Fusion.Adv Ther (Weinh). 2021 May;4(5):2000224. doi: 10.1002/adtp.202000224. Epub 2021 Feb 22. Adv Ther (Weinh). 2021. PMID: 33786369 Free PMC article.
-
Comorbidities' potential impacts on severe and non-severe patients with COVID-19: A systematic review and meta-analysis.Medicine (Baltimore). 2021 Mar 26;100(12):e24971. doi: 10.1097/MD.0000000000024971. Medicine (Baltimore). 2021. PMID: 33761654 Free PMC article.
-
Mesenchymal stromal cells as potential immunomodulatory players in severe acute respiratory distress syndrome induced by SARS-CoV-2 infection.World J Stem Cells. 2020 Aug 26;12(8):731-751. doi: 10.4252/wjsc.v12.i8.731. World J Stem Cells. 2020. PMID: 32952855 Free PMC article. Review.
-
Conventional and Microfluidic Methods for the Detection of Nucleic Acid of SARS-CoV-2.Micromachines (Basel). 2022 Apr 17;13(4):636. doi: 10.3390/mi13040636. Micromachines (Basel). 2022. PMID: 35457940 Free PMC article. Review.
-
Current Advances in Paper-Based Biosensor Technologies for Rapid COVID-19 Diagnosis.Biochip J. 2022;16(4):376-396. doi: 10.1007/s13206-022-00078-9. Epub 2022 Aug 10. Biochip J. 2022. PMID: 35968255 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
