Genome-wide siRNA screening reveals that DCAF4-mediated ubiquitination of optineurin stimulates autophagic degradation of Cu,Zn-superoxide dismutase
- PMID: 32014991
- PMCID: PMC7062173
- DOI: 10.1074/jbc.RA119.010239
Genome-wide siRNA screening reveals that DCAF4-mediated ubiquitination of optineurin stimulates autophagic degradation of Cu,Zn-superoxide dismutase
Abstract
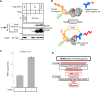
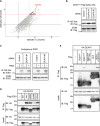
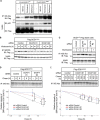
Cu, Zn superoxide dismutase (SOD1) is one of the genes implicated in the devastating neurodegenerative disorder amyotrophic lateral sclerosis (ALS). Although the precise mechanisms of SOD1 mutant (SOD1mut)-induced motoneuron toxicity are still unclear, defects in SOD1 proteostasis are known to have a critical role in ALS pathogenesis. We previously reported that the SOD1mut adopts a conformation that exposes a Derlin-1-binding region (DBR) and that DBR-exposed SOD1 interacts with Derlin-1, leading to motoneuron death. We also found that an environmental change, i.e. zinc depletion, induces a conformational change in WT SOD1 (SOD1WT) to the DBR-exposed conformation, suggesting the presence of an equilibrium state between the DBR-masked and DBR-exposed states even with SOD1WT Here, we conducted a high-throughput screening based on time-resolved FRET to further investigate the SOD1WT conformational change, and we used a genome-wide siRNA screen to search for regulators of SOD1 proteostasis. This screen yielded 30 candidate genes that maintained an absence of the DBR-exposed SOD1WT conformation. Among these genes was one encoding DDB1- and CUL4-associated factor 4 (DCAF4), a substrate receptor of the E3 ubiquitin-protein ligase complex. Of note, we found that DCAF4 mediates the ubiquitination of an ALS-associated protein and autophagy receptor, optineurin (OPTN), and facilitates autophagic degradation of DBR-exposed SOD1. In summary, our screen identifies DCAF4 as being required for proper proteostasis of DBR-exposed SOD1, which may have potential relevance for the development of therapies for managing ALS.
Keywords: DDB1 and CUL4 associated factor 4 (DCAF4); amyotrophic lateral sclerosis (ALS) (Lou Gehrig disease); autophagy; genome-wide siRNA screen; protein aggregation; protein misfolding; proteostasis; superoxide dismutase 1 (SOD1).
© 2020 Homma et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


Similar articles
-
TNF receptor-associated factor 6 interacts with ALS-linked misfolded superoxide dismutase 1 and promotes aggregation.J Biol Chem. 2020 Mar 20;295(12):3808-3825. doi: 10.1074/jbc.RA119.011215. Epub 2020 Feb 6. J Biol Chem. 2020. PMID: 32029478 Free PMC article.
-
ALS-Related Mutant SOD1 Aggregates Interfere with Mitophagy by Sequestering the Autophagy Receptor Optineurin.Int J Mol Sci. 2020 Oct 13;21(20):7525. doi: 10.3390/ijms21207525. Int J Mol Sci. 2020. PMID: 33065963 Free PMC article.
-
A systematic immunoprecipitation approach reinforces the concept of common conformational alterations in amyotrophic lateral sclerosis-linked SOD1 mutants.Neurobiol Dis. 2015 Oct;82:478-486. doi: 10.1016/j.nbd.2015.08.010. Epub 2015 Aug 18. Neurobiol Dis. 2015. PMID: 26297318
-
SOD1 in neurotoxicity and its controversial roles in SOD1 mutation-negative ALS.Adv Biol Regul. 2016 Jan;60:95-104. doi: 10.1016/j.jbior.2015.10.006. Epub 2015 Oct 31. Adv Biol Regul. 2016. PMID: 26563614 Review.
-
[Prion-like Properties of Misfolded Cu/Zn-superoxide Dismutase in Amyotrophic Lateral Sclerosis: Update and Perspectives].Yakugaku Zasshi. 2019;139(7):1015-1019. doi: 10.1248/yakushi.18-00165-5. Yakugaku Zasshi. 2019. PMID: 31257248 Review. Japanese.
Cited by
-
SoDCoD: a comprehensive database of Cu/Zn superoxide dismutase conformational diversity caused by ALS-linked gene mutations and other perturbations.Database (Oxford). 2024 Aug 8;2024:0. doi: 10.1093/database/baae064. Database (Oxford). 2024. PMID: 39126203 Free PMC article.
-
Ubiquitin signaling in neurodegenerative diseases: an autophagy and proteasome perspective.Cell Death Differ. 2021 Feb;28(2):439-454. doi: 10.1038/s41418-020-00667-x. Epub 2020 Nov 18. Cell Death Differ. 2021. PMID: 33208890 Free PMC article. Review.
References
-
- Reaume A. G., Elliott J. L., Hoffman E. K., Kowall N. W., Ferrante R. J., Siwek D. F., Wilcox H. M., Flood D. G., Beal M. F., Brown R. H. Jr., Scott R. W., and Snider W. D. (1996) Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat. Genet. 13, 43–47 10.1038/ng0596-43 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

