Different human resting memory CD4+ T cell subsets show similar low inducibility of latent HIV-1 proviruses
- PMID: 31996465
- PMCID: PMC7875249
- DOI: 10.1126/scitranslmed.aax6795
Different human resting memory CD4+ T cell subsets show similar low inducibility of latent HIV-1 proviruses
Abstract
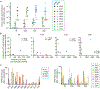
The latent reservoir of HIV-1 in resting CD4+ T cells is a major barrier to cure. It is unclear whether the latent reservoir resides principally in particular subsets of CD4+ T cells, a finding that would have implications for understanding its stability and developing curative therapies. Recent work has shown that proliferation of HIV-1-infected CD4+ T cells is a major factor in the generation and persistence of the latent reservoir and that latently infected T cells that have clonally expanded in vivo can proliferate in vitro without producing virions. In certain CD4+ memory T cell subsets, the provirus may be in a deeper state of latency, allowing the cell to proliferate without producing viral proteins, thus permitting escape from immune clearance. To evaluate this possibility, we used a multiple stimulation viral outgrowth assay to culture resting naïve, central memory (TCM), transitional memory (TTM), and effector memory (TEM) CD4+ T cells from 10 HIV-1-infected individuals on antiretroviral therapy. On average, only 1.7% of intact proviruses across all T cell subsets were induced to transcribe viral genes and release replication-competent virus after stimulation of the cells. We found no consistent enrichment of intact or inducible proviruses in any T cell subset. Furthermore, we observed notable plasticity among the canonical memory T cell subsets after activation in vitro and saw substantial person-to-person variability in the inducibility of infectious virus release. This finding complicates the vision for a targeted approach for HIV-1 cure based on T cell memory subsets.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures

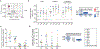
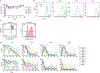
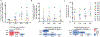


Similar articles
-
Atlas of the HIV-1 Reservoir in Peripheral CD4 T Cells of Individuals on Successful Antiretroviral Therapy.mBio. 2021 Dec 21;12(6):e0307821. doi: 10.1128/mBio.03078-21. Epub 2021 Nov 30. mBio. 2021. PMID: 34844430 Free PMC article.
-
HIV-1 Genomes Are Enriched in Memory CD4+ T-Cells with Short Half-Lives.mBio. 2021 Oct 26;12(5):e0244721. doi: 10.1128/mBio.02447-21. Epub 2021 Sep 21. mBio. 2021. PMID: 34544282 Free PMC article.
-
Genetic Diversity, Compartmentalization, and Age of HIV Proviruses Persisting in CD4+ T Cell Subsets during Long-Term Combination Antiretroviral Therapy.J Virol. 2020 Feb 14;94(5):e01786-19. doi: 10.1128/JVI.01786-19. Print 2020 Feb 14. J Virol. 2020. PMID: 31776273 Free PMC article.
-
Low Inducibility of Latent Human Immunodeficiency Virus Type 1 Proviruses as a Major Barrier to Cure.J Infect Dis. 2021 Feb 15;223(12 Suppl 2):13-21. doi: 10.1093/infdis/jiaa649. J Infect Dis. 2021. PMID: 33586775 Free PMC article. Review.
-
Targeting the Latent Reservoir for HIV-1.Immunity. 2018 May 15;48(5):872-895. doi: 10.1016/j.immuni.2018.04.030. Immunity. 2018. PMID: 29768175 Free PMC article. Review.
Cited by
-
New Assay Reveals Vast Excess of Defective over Intact HIV-1 Transcripts in Antiretroviral Therapy-Suppressed Individuals.J Virol. 2022 Dec 21;96(24):e0160522. doi: 10.1128/jvi.01605-22. Epub 2022 Nov 30. J Virol. 2022. PMID: 36448806 Free PMC article.
-
IRF7 expression correlates with HIV latency reversal upon specific blockade of immune activation.Front Immunol. 2022 Sep 5;13:1001068. doi: 10.3389/fimmu.2022.1001068. eCollection 2022. Front Immunol. 2022. PMID: 36131914 Free PMC article.
-
HIV post-treatment controllers have distinct immunological and virological features.Proc Natl Acad Sci U S A. 2023 Mar 14;120(11):e2218960120. doi: 10.1073/pnas.2218960120. Epub 2023 Mar 6. Proc Natl Acad Sci U S A. 2023. PMID: 36877848 Free PMC article.
-
Clearance of HIV-1 or SIV reservoirs by promotion of apoptosis and inhibition of autophagy: Targeting intracellular molecules in cure-directed strategies.J Leukoc Biol. 2022 Nov;112(5):1245-1259. doi: 10.1002/JLB.4MR0222-606. Epub 2022 Mar 31. J Leukoc Biol. 2022. PMID: 35362118 Free PMC article. Review.
-
Spontaneous HIV expression during suppressive ART is associated with the magnitude and function of HIV-specific CD4+ and CD8+ T cells.Cell Host Microbe. 2023 Sep 13;31(9):1507-1522.e5. doi: 10.1016/j.chom.2023.08.006. Cell Host Microbe. 2023. PMID: 37708853 Free PMC article.
References
-
- Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF, Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells, Nat. Med 9, 727–728 (2003). - PubMed
-
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF, Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy, Nat. Med 5, 512–517 (1999). - PubMed
-
- Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF, In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency, Nat. Med 1, 1284–1290 (1995). - PubMed
-
- Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF, Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection, Nature 387, 183–188 (1997). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

