Burkholderia pseudomallei invades the olfactory nerve and bulb after epithelial injury in mice and causes the formation of multinucleated giant glial cells in vitro
- PMID: 31978058
- PMCID: PMC7002012
- DOI: 10.1371/journal.pntd.0008017
Burkholderia pseudomallei invades the olfactory nerve and bulb after epithelial injury in mice and causes the formation of multinucleated giant glial cells in vitro
Abstract
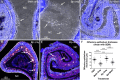
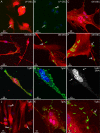
The infectious disease melioidosis is caused by the bacterium Burkholderia pseudomallei. Melioidosis is characterised by high mortality and morbidity and can involve the central nervous system (CNS). We have previously discovered that B. pseudomallei can infect the CNS via the olfactory and trigeminal nerves in mice. We have shown that the nerve path is dependent on mouse strain, with outbred mice showing resistance to olfactory nerve infection. Damage to the nasal epithelium by environmental factors is common, and we hypothesised that injury to the olfactory epithelium may increase the vulnerability of the olfactory nerve to microbial insult. We therefore investigated this, using outbred mice that were intranasally inoculated with B. pseudomallei, with or without methimazole-induced injury to the olfactory neuroepithelium. Methimazole-mediated injury resulted in increased B. pseudomallei invasion of the olfactory epithelium, and only in pre-injured animals were bacteria found in the olfactory nerve and bulb. In vitro assays demonstrated that B. pseudomallei readily infected glial cells isolated from the olfactory and trigeminal nerves (olfactory ensheathing cells and trigeminal Schwann cells, respectively). Bacteria were degraded by some cells but persisted in other cells, which led to the formation of multinucleated giant cells (MNGCs), with olfactory ensheathing cells less likely to form MNGCs than Schwann cells. Double Cap mutant bacteria, lacking the protein BimA, did not form MNGCs. These data suggest that injuries to the olfactory epithelium expose the primary olfactory nervous system to bacterial invasion, which can then result in CNS infection with potential pathogenic consequences for the glial cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Burkholderia pseudomallei penetrates the brain via destruction of the olfactory and trigeminal nerves: implications for the pathogenesis of neurological melioidosis.mBio. 2014 Apr 15;5(2):e00025. doi: 10.1128/mBio.00025-14. mBio. 2014. PMID: 24736221 Free PMC article.
-
Novel insights into the glia limitans of the olfactory nervous system.J Comp Neurol. 2019 May 1;527(7):1228-1244. doi: 10.1002/cne.24618. Epub 2019 Jan 16. J Comp Neurol. 2019. PMID: 30592044
-
Burkholderia pseudomallei Rapidly Infects the Brain Stem and Spinal Cord via the Trigeminal Nerve after Intranasal Inoculation.Infect Immun. 2016 Aug 19;84(9):2681-8. doi: 10.1128/IAI.00361-16. Print 2016 Sep. Infect Immun. 2016. PMID: 27382023 Free PMC article.
-
Pathogenicity and virulence of Burkholderia pseudomallei.Virulence. 2022 Dec;13(1):1945-1965. doi: 10.1080/21505594.2022.2139063. Virulence. 2022. PMID: 36271712 Free PMC article. Review.
-
Pathogens penetrating the central nervous system: infection pathways and the cellular and molecular mechanisms of invasion.Clin Microbiol Rev. 2014 Oct;27(4):691-726. doi: 10.1128/CMR.00118-13. Clin Microbiol Rev. 2014. PMID: 25278572 Free PMC article. Review.
Cited by
-
Chlamydia pneumoniae can infect the central nervous system via the olfactory and trigeminal nerves and contributes to Alzheimer's disease risk.Sci Rep. 2022 Feb 17;12(1):2759. doi: 10.1038/s41598-022-06749-9. Sci Rep. 2022. PMID: 35177758 Free PMC article.
-
Cycle-Inhibiting Factor Is Associated with Burkholderia pseudomallei Invasion in Human Neuronal Cells.Biology (Basel). 2022 Oct 1;11(10):1439. doi: 10.3390/biology11101439. Biology (Basel). 2022. PMID: 36290346 Free PMC article.
-
In Vitro Roles of Burkholderia Intracellular Motility A (BimA) in Infection of Human Neuroblastoma Cell Line.Microbiol Spectr. 2023 Aug 17;11(4):e0132023. doi: 10.1128/spectrum.01320-23. Epub 2023 Jul 6. Microbiol Spectr. 2023. PMID: 37409935 Free PMC article.
-
Virulence of Burkholderia pseudomallei ATS2021 Unintentionally Imported to United States in Aromatherapy Spray.Emerg Infect Dis. 2024 Oct;30(10):2056-2069. doi: 10.3201/eid3010.240084. Emerg Infect Dis. 2024. PMID: 39320153 Free PMC article.
-
Type VI Secretion System Accessory Protein TagAB-5 Promotes Burkholderia pseudomallei Pathogenicity in Human Microglia.Biomedicines. 2023 Oct 30;11(11):2927. doi: 10.3390/biomedicines11112927. Biomedicines. 2023. PMID: 38001928 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

