Single cell analysis reveals human cytomegalovirus drives latently infected cells towards an anergic-like monocyte state
- PMID: 31967545
- PMCID: PMC7039680
- DOI: 10.7554/eLife.52168
Single cell analysis reveals human cytomegalovirus drives latently infected cells towards an anergic-like monocyte state
Abstract
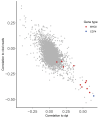
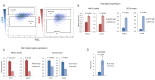
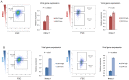
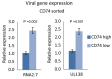
Human cytomegalovirus (HCMV) causes a lifelong infection through establishment of latency. Although reactivation from latency can cause life-threatening disease, our molecular understanding of HCMV latency is incomplete. Here we use single cell RNA-seq analysis to characterize latency in monocytes and hematopoietic stem and progenitor cells (HSPCs). In monocytes, we identify host cell surface markers that enable enrichment of latent cells harboring higher viral transcript levels, which can reactivate more efficiently, and are characterized by reduced intrinsic immune response that is important for viral gene expression. Significantly, in latent HSPCs, viral transcripts could be detected only in monocyte progenitors and were also associated with reduced immune-response. Overall, our work indicates that regardless of the developmental stage in which HCMV infects, HCMV drives hematopoietic cells towards a weaker immune-responsive monocyte state and that this anergic-like state is crucial for the virus ability to express its transcripts and to eventually reactivate.
Keywords: cytomegalovirus; hematopoietic stem and progenitor cells; herpesvirus; human; infectious disease; latency; microbiology; reactivation; single-cell RNA-seq.
Plain language summary
Most people around the world unknowingly carry the human cytomegalovirus, as this virus can become dormant after infection and hide in small numbers of blood stem cells (which give rise to blood and immune cells). Dormant viruses still make their host cells read their genetic information and create viral proteins – a process known as gene expression – but they do not use them to quickly multiply. However, it is possible for the cytomegalovirus to reawaken at a later stage and start replicating again, which can be fatal for people with weakened immune systems. It is therefore important to understand exactly how the virus can stay dormant, and how it reactivates. Only certain infected cells allow dormant viruses to later reactivate; in others, it never starts to multiply again. Techniques that can monitor individual cells are therefore needed to understand how the host cells and the viruses interact during dormant infection and reactivation. To investigate this, Shnayder et al. infected blood stem cells in the laboratory and used a method known as single-cell RNA analysis, which highlights all the genes (including viral genes) that are expressed in a cell. This showed that in certain cells, the virus dampens the cell defenses, leading to a higher rate of viral gene expression and, in turn, easier reactivation. Further experiments showed that the blood stem cells that expressed the viral genes were marked to become a type of immune cells known as monocytes. In turn, these infected monocytes were shown to be less able to defend the body against infection, suggesting that latent human cytomegalovirus suppresses the body’s innate immune response. The reactivation of human cytomegalovirus is a dangerous issue for patients who have just received an organ or blood stem cells transplant. The study by Shnayder et al. indicates that treatments that boost innate immunity may help to prevent the virus from reawakening, but more work is needed to test this theory.
© 2020, Shnayder et al.
Conflict of interest statement
MS, AN, BR, BB, ML, NF, EP, SA, EB, DG, AA, BS, JS, NS, MS No competing interests declared
Figures






Similar articles
-
Human cytomegalovirus modulates monocyte-mediated innate immune responses during short-term experimental latency in vitro.J Virol. 2014 Aug;88(16):9391-405. doi: 10.1128/JVI.00934-14. Epub 2014 Jun 11. J Virol. 2014. PMID: 24920803 Free PMC article.
-
Human cytomegalovirus reprogrammes haematopoietic progenitor cells into immunosuppressive monocytes to achieve latency.Nat Microbiol. 2018 Apr;3(4):503-513. doi: 10.1038/s41564-018-0131-9. Epub 2018 Mar 27. Nat Microbiol. 2018. PMID: 29588542 Free PMC article.
-
Latency-Associated Expression of Human Cytomegalovirus US28 Attenuates Cell Signaling Pathways To Maintain Latent Infection.mBio. 2017 Dec 5;8(6):e01754-17. doi: 10.1128/mBio.01754-17. mBio. 2017. PMID: 29208743 Free PMC article.
-
Human cytomegalovirus infection of human hematopoietic progenitor cells.Leuk Lymphoma. 1999 Mar;33(1-2):1-13. doi: 10.3109/10428199909093720. Leuk Lymphoma. 1999. PMID: 10194116 Review.
-
Advances in Model Systems for Human Cytomegalovirus Latency and Reactivation.mBio. 2022 Feb 22;13(1):e0172421. doi: 10.1128/mbio.01724-21. Epub 2022 Jan 11. mBio. 2022. PMID: 35012351 Free PMC article. Review.
Cited by
-
Human cytomegalovirus and neonatal infection.Curr Res Microb Sci. 2024 Jun 24;7:100257. doi: 10.1016/j.crmicr.2024.100257. eCollection 2024. Curr Res Microb Sci. 2024. PMID: 39070527 Free PMC article. Review.
-
Virally encoded interleukin-6 facilitates KSHV replication in monocytes and induction of dysfunctional macrophages.PLoS Pathog. 2023 Oct 26;19(10):e1011703. doi: 10.1371/journal.ppat.1011703. eCollection 2023 Oct. PLoS Pathog. 2023. PMID: 37883374 Free PMC article.
-
Single-cell transcriptomics identifies Gadd45b as a regulator of herpesvirus-reactivating neurons.EMBO Rep. 2022 Feb 3;23(2):e53543. doi: 10.15252/embr.202153543. Epub 2021 Nov 29. EMBO Rep. 2022. PMID: 34842321 Free PMC article.
-
Human Cytomegalovirus Host Interactions: EGFR and Host Cell Signaling Is a Point of Convergence Between Viral Infection and Functional Changes in Infected Cells.Front Microbiol. 2021 May 7;12:660901. doi: 10.3389/fmicb.2021.660901. eCollection 2021. Front Microbiol. 2021. PMID: 34025614 Free PMC article. Review.
-
Functional and molecular dissection of HCMV long non-coding RNAs.Sci Rep. 2022 Nov 11;12(1):19303. doi: 10.1038/s41598-022-23317-3. Sci Rep. 2022. PMID: 36369338 Free PMC article.
References
-
- Avdic S, Cao JZ, McSharry BP, Clancy LE, Brown R, Steain M, Gottlieb DJ, Abendroth A, Slobedman B. Human Cytomegalovirus interleukin-10 polarizes monocytes toward a deactivated M2c phenotype to repress host immune responses. Journal of Virology. 2013;87:10273–10282. doi: 10.1128/JVI.00912-13. - DOI - PMC - PubMed
-
- Blood Atlas 2019. VTCN1 the human protein atlas. The Human Protein Atlas. https://www.proteinatlas.org/ENSG00000134258-VTCN1/blood
Publication types
MeSH terms
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

