Bacterial Hsp70 resolves misfolded states and accelerates productive folding of a multi-domain protein
- PMID: 31953415
- PMCID: PMC6969021
- DOI: 10.1038/s41467-019-14245-4
Bacterial Hsp70 resolves misfolded states and accelerates productive folding of a multi-domain protein
Abstract
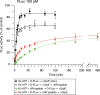
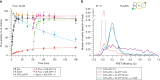
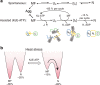
The ATP-dependent Hsp70 chaperones (DnaK in E. coli) mediate protein folding in cooperation with J proteins and nucleotide exchange factors (E. coli DnaJ and GrpE, respectively). The Hsp70 system prevents protein aggregation and increases folding yields. Whether it also enhances the rate of folding remains unclear. Here we show that DnaK/DnaJ/GrpE accelerate the folding of the multi-domain protein firefly luciferase (FLuc) ~20-fold over the rate of spontaneous folding measured in the absence of aggregation. Analysis by single-pair FRET and hydrogen/deuterium exchange identified inter-domain misfolding as the cause of slow folding. DnaK binding expands the misfolded region and thereby resolves the kinetically-trapped intermediates, with folding occurring upon GrpE-mediated release. In each round of release DnaK commits a fraction of FLuc to fast folding, circumventing misfolding. We suggest that by resolving misfolding and accelerating productive folding, the bacterial Hsp70 system can maintain proteins in their native states under otherwise denaturing stress conditions.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Probing the different chaperone activities of the bacterial HSP70-HSP40 system using a thermolabile luciferase substrate.Proteins. 2011 Jun;79(6):1991-8. doi: 10.1002/prot.23024. Epub 2011 Apr 12. Proteins. 2011. PMID: 21488102
-
Regulation of ATPase and chaperone cycle of DnaK from Thermus thermophilus by the nucleotide exchange factor GrpE.J Mol Biol. 2001 Feb 2;305(5):1173-83. doi: 10.1006/jmbi.2000.4373. J Mol Biol. 2001. PMID: 11162122
-
Action of the Hsp70 chaperone system observed with single proteins.Nat Commun. 2015 Feb 17;6:6307. doi: 10.1038/ncomms7307. Nat Commun. 2015. PMID: 25686738
-
Interferon-gamma is a target for binding and folding by both Escherichia coli chaperone model systems GroEL/GroES and DnaK/DnaJ/GrpE.Biochimie. 1998 Aug-Sep;80(8-9):729-37. doi: 10.1016/s0300-9084(99)80026-1. Biochimie. 1998. PMID: 9865495 Review.
-
Substrate Interaction Networks of the Escherichia coli Chaperones: Trigger Factor, DnaK and GroEL.Adv Exp Med Biol. 2015;883:271-94. doi: 10.1007/978-3-319-23603-2_15. Adv Exp Med Biol. 2015. PMID: 26621473 Review.
Cited by
-
Non-Equilibrium Protein Folding and Activation by ATP-Driven Chaperones.Biomolecules. 2022 Jun 15;12(6):832. doi: 10.3390/biom12060832. Biomolecules. 2022. PMID: 35740957 Free PMC article.
-
J-domain protein chaperone circuits in proteostasis and disease.Trends Cell Biol. 2023 Jan;33(1):30-47. doi: 10.1016/j.tcb.2022.05.004. Epub 2022 Jun 18. Trends Cell Biol. 2023. PMID: 35729039 Free PMC article. Review.
-
Molecular dissection of amyloid disaggregation by human HSP70.Nature. 2020 Nov;587(7834):483-488. doi: 10.1038/s41586-020-2904-6. Epub 2020 Nov 11. Nature. 2020. PMID: 33177717
-
Real-time single-molecule observation of chaperone-assisted protein folding.Sci Adv. 2022 Dec 14;8(50):eadd0922. doi: 10.1126/sciadv.add0922. Epub 2022 Dec 14. Sci Adv. 2022. PMID: 36516244 Free PMC article.
-
Single-molecule evidence of Entropic Pulling by Hsp70 chaperones.Nat Commun. 2024 Oct 8;15(1):8604. doi: 10.1038/s41467-024-52674-y. Nat Commun. 2024. PMID: 39379347 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

