Monocytes and Monocyte-Derived Antigen-Presenting Cells Have Distinct Gene Signatures in Experimental Model of Multiple Sclerosis
- PMID: 31849962
- PMCID: PMC6889845
- DOI: 10.3389/fimmu.2019.02779
Monocytes and Monocyte-Derived Antigen-Presenting Cells Have Distinct Gene Signatures in Experimental Model of Multiple Sclerosis
Abstract
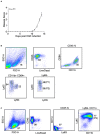
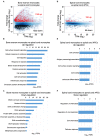
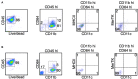
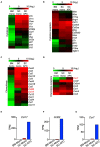
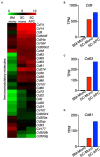
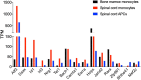
Multiple sclerosis (MS) is a chronic inflammatory disease mediated by a complex interaction between the autoreactive lymphocytes and the effector myeloid cells within the central nervous system (CNS). In a murine model of MS, experimental autoimmune encephalomyelitis (EAE), Ly6Chi monocytes migrate into the CNS and further differentiate into antigen-presenting cells (APCs) during disease progression. Currently, there is no information about gene signatures that can distinguish between monocytes and the monocyte-derived APCs. We developed a surface marker-based strategy to distinguish between these two cell types during the stage of EAE when the clinical symptoms were most severe, and performed transcriptome analysis to compare their gene expression. We report here that the inflammatory CNS environment substantially alters gene expression of monocytes, compared to the monocyte differentiation process within CNS. Monocytes in the CNS express genes that encode proinflammatory cytokines and chemokines, and their expression is mostly maintained when the cells differentiate. Moreover, monocyte-derived APCs express surface markers associated with both dendritic cells and macrophages, and have a significant up-regulation of genes that are critical for antigen presentation. Furthermore, we found that Ccl17, Ccl22, and Ccr7 are expressed in monocyte-derived APCs but not the Ly6Chi monocytes. These findings may shed light on identifying molecular signals that control monocyte differentiation and functions during EAE.
Keywords: RNA-Seq; antigen-presenting cells; experimental autoimmune encephalomyelitis; monocytes; multiple sclerosis.
Copyright © 2019 Monaghan, Zheng, Hu and Wan.
Figures



Similar articles
-
Sex-Specific Effects of Microglia-Like Cell Engraftment during Experimental Autoimmune Encephalomyelitis.Int J Mol Sci. 2020 Sep 17;21(18):6824. doi: 10.3390/ijms21186824. Int J Mol Sci. 2020. PMID: 32957621 Free PMC article.
-
Antigen-oriented T cell migration contributes to myelin peptide induced-EAE and immune tolerance.Clin Immunol. 2016 Aug;169:36-46. doi: 10.1016/j.clim.2016.06.004. Epub 2016 Jun 17. Clin Immunol. 2016. PMID: 27327113
-
TRPM2 Exacerbates Central Nervous System Inflammation in Experimental Autoimmune Encephalomyelitis by Increasing Production of CXCL2 Chemokines.J Neurosci. 2018 Sep 26;38(39):8484-8495. doi: 10.1523/JNEUROSCI.2203-17.2018. Epub 2018 Sep 10. J Neurosci. 2018. PMID: 30201769 Free PMC article.
-
The critical role of antigen-presentation-induced cytokine crosstalk in the central nervous system in multiple sclerosis and experimental autoimmune encephalomyelitis.J Interferon Cytokine Res. 2011 Oct;31(10):753-68. doi: 10.1089/jir.2011.0052. Epub 2011 Sep 15. J Interferon Cytokine Res. 2011. PMID: 21919736 Free PMC article. Review.
-
The role of antigen presenting cells in multiple sclerosis.Biochim Biophys Acta. 2011 Feb;1812(2):265-74. doi: 10.1016/j.bbadis.2010.07.008. Epub 2010 Jul 15. Biochim Biophys Acta. 2011. PMID: 20637861 Free PMC article. Review.
Cited by
-
Fc multimers effectively treat murine models of multiple sclerosis.Front Immunol. 2023 Aug 11;14:1199747. doi: 10.3389/fimmu.2023.1199747. eCollection 2023. Front Immunol. 2023. PMID: 37638040 Free PMC article.
-
Tetramerization of STAT5 regulates monocyte differentiation and the dextran sulfate sodium-induced colitis in mice.Front Immunol. 2023 Apr 20;14:1117828. doi: 10.3389/fimmu.2023.1117828. eCollection 2023. Front Immunol. 2023. PMID: 37153611 Free PMC article.
-
A miRNA/CXCR4 signaling axis impairs monopoiesis and angiogenesis in diabetic critical limb ischemia.JCI Insight. 2023 Apr 10;8(7):e163360. doi: 10.1172/jci.insight.163360. JCI Insight. 2023. PMID: 36821386 Free PMC article.
-
Tetramerization of STAT5 promotes autoimmune-mediated neuroinflammation.Proc Natl Acad Sci U S A. 2021 Dec 28;118(52):e2116256118. doi: 10.1073/pnas.2116256118. Proc Natl Acad Sci U S A. 2021. PMID: 34934004 Free PMC article.
-
Landscape of Hopx expression in cells of the immune system.Heliyon. 2021 Nov 2;7(11):e08311. doi: 10.1016/j.heliyon.2021.e08311. eCollection 2021 Nov. Heliyon. 2021. PMID: 34805566 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

