CD19-targeting CAR T cell immunotherapy outcomes correlate with genomic modification by vector integration
- PMID: 31845905
- PMCID: PMC6994131
- DOI: 10.1172/JCI130144
CD19-targeting CAR T cell immunotherapy outcomes correlate with genomic modification by vector integration
Abstract
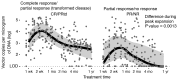
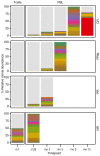
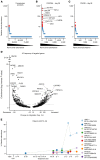
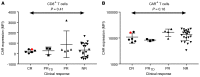
Chimeric antigen receptor-engineered T cells targeting CD19 (CART19) provide an effective treatment for pediatric acute lymphoblastic leukemia but are less effective for chronic lymphocytic leukemia (CLL), focusing attention on improving efficacy. CART19 harbor an engineered receptor, which is delivered through lentiviral vector integration, thereby marking cell lineages and modifying the cellular genome by insertional mutagenesis. We recently reported that vector integration within the host TET2 gene was associated with CLL remission. Here, we investigated clonal population structure and therapeutic outcomes in another 39 patients by high-throughput sequencing of vector-integration sites. Genes at integration sites enriched in responders were commonly found in cell-signaling and chromatin modification pathways, suggesting that insertional mutagenesis in these genes promoted therapeutic T cell proliferation. We also developed a multivariate model based on integration-site distributions and found that data from preinfusion products forecasted response in CLL successfully in discovery and validation cohorts and, in day 28 samples, reported responders to CLL therapy with high accuracy. These data clarify how insertional mutagenesis can modulate cell proliferation in CART19 therapy and how data on integration-site distributions can be linked to treatment outcomes.
Keywords: Immunotherapy; Microbiology.
Conflict of interest statement
Figures

Similar articles
-
CAR T Cell Therapy in Acute Lymphoblastic Leukemia and Potential for Chronic Lymphocytic Leukemia.Curr Treat Options Oncol. 2016 Jun;17(6):28. doi: 10.1007/s11864-016-0406-4. Curr Treat Options Oncol. 2016. PMID: 27098534 Review.
-
CD19 Chimeric Antigen Receptor T Cells From Patients With Chronic Lymphocytic Leukemia Display an Elevated IFN-γ Production Profile.J Immunother. 2018 Feb/Mar;41(2):73-83. doi: 10.1097/CJI.0000000000000193. J Immunother. 2018. PMID: 29315094 Free PMC article.
-
Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells.Nature. 2018 Jun;558(7709):307-312. doi: 10.1038/s41586-018-0178-z. Epub 2018 May 30. Nature. 2018. PMID: 29849141 Free PMC article.
-
Ibrutinib for improved chimeric antigen receptor T-cell production for chronic lymphocytic leukemia patients.Int J Cancer. 2021 Jan 15;148(2):419-428. doi: 10.1002/ijc.33212. Epub 2020 Jul 28. Int J Cancer. 2021. PMID: 32683672
-
CD19-Chimeric Antigen Receptor T Cells for Treatment of Chronic Lymphocytic Leukaemia and Acute Lymphoblastic Leukaemia.Scand J Immunol. 2015 Oct;82(4):307-19. doi: 10.1111/sji.12331. Scand J Immunol. 2015. PMID: 26099639 Review.
Cited by
-
CAR T Cell Therapy for Solid Tumors: Bright Future or Dark Reality?Mol Ther. 2020 Nov 4;28(11):2320-2339. doi: 10.1016/j.ymthe.2020.09.015. Epub 2020 Sep 16. Mol Ther. 2020. PMID: 32979309 Free PMC article. Review.
-
Choosing the Right Tool for Genetic Engineering: Clinical Lessons from Chimeric Antigen Receptor-T Cells.Hum Gene Ther. 2021 Oct;32(19-20):1044-1058. doi: 10.1089/hum.2021.173. Hum Gene Ther. 2021. PMID: 34662233 Free PMC article. Review.
-
Epigenetic Profiling and Response to CD19 Chimeric Antigen Receptor T-Cell Therapy in B-Cell Malignancies.J Natl Cancer Inst. 2022 Mar 8;114(3):436-445. doi: 10.1093/jnci/djab194. J Natl Cancer Inst. 2022. PMID: 34581788 Free PMC article.
-
Sleeping Beauty-engineered CAR T cells achieve antileukemic activity without severe toxicities.J Clin Invest. 2020 Nov 2;130(11):6021-6033. doi: 10.1172/JCI138473. J Clin Invest. 2020. PMID: 32780725 Free PMC article. Clinical Trial.
-
Context-aware synthetic biology by controller design: Engineering the mammalian cell.Cell Syst. 2021 Jun 16;12(6):561-592. doi: 10.1016/j.cels.2021.05.011. Cell Syst. 2021. PMID: 34139166 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

