Epigenomic analysis of gastrulation identifies a unique chromatin state for primed pluripotency
- PMID: 31844322
- PMCID: PMC7362285
- DOI: 10.1038/s41588-019-0545-1
Epigenomic analysis of gastrulation identifies a unique chromatin state for primed pluripotency
Abstract
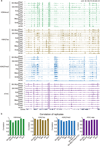
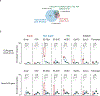
Around implantation, the epiblast (Epi) transits from naïve to primed pluripotency, before giving rise to the three germ layers. How chromatin is reconfigured during this developmental window remains poorly understood. We performed a genome-wide investigation of chromatin landscapes during this period. We find that enhancers in ectoderm are already pre-accessible in embryonic day 6.5 (E6.5) Epi when cells enter a primed pluripotent state. Unexpectedly, strong trimethylation of histone H3 at lysine 4 (H3K4me3) emerges at developmental gene promoters in E6.5 Epi and positively correlates with H3K27me3, thus establishing bivalency. These genes also show enhanced spatial interactions. Both the strong bivalency and spatial clustering are virtually absent in preimplantation embryos and are markedly reduced in fate-committed lineages. Finally, we show that KMT2B is essential for establishing bivalent H3K4me3 at E6.5 but becomes partially dispensable later. Its deficiency leads to impaired activation of developmental genes and subsequent embryonic lethality. Thus, our data characterize lineage-specific chromatin reconfiguration and a unique chromatin state for primed pluripotency.
Figures




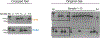
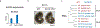








Similar articles
-
A unique chromatin signature uncovers early developmental enhancers in humans.Nature. 2011 Feb 10;470(7333):279-83. doi: 10.1038/nature09692. Epub 2010 Dec 15. Nature. 2011. PMID: 21160473 Free PMC article.
-
Derivation of novel human ground state naive pluripotent stem cells.Nature. 2013 Dec 12;504(7479):282-6. doi: 10.1038/nature12745. Epub 2013 Oct 30. Nature. 2013. PMID: 24172903
-
Genome-Wide Studies Reveal that H3K4me3 Modification in Bivalent Genes Is Dynamically Regulated during the Pluripotent Cell Cycle and Stabilized upon Differentiation.Mol Cell Biol. 2015 Dec 7;36(4):615-27. doi: 10.1128/MCB.00877-15. Print 2016 Feb 15. Mol Cell Biol. 2015. PMID: 26644406 Free PMC article.
-
Chromatin remodeling and bivalent histone modifications in embryonic stem cells.EMBO Rep. 2015 Dec;16(12):1609-19. doi: 10.15252/embr.201541011. Epub 2015 Nov 9. EMBO Rep. 2015. PMID: 26553936 Free PMC article. Review.
-
Epigenetic modulation by TFII-I during embryonic stem cell differentiation.J Cell Biochem. 2012 Oct;113(10):3056-60. doi: 10.1002/jcb.24202. J Cell Biochem. 2012. PMID: 22628223 Review.
Cited by
-
Comprehensive chromatin proteomics resolves functional phases of pluripotency and identifies changes in regulatory components.Nucleic Acids Res. 2023 Apr 11;51(6):2671-2690. doi: 10.1093/nar/gkad058. Nucleic Acids Res. 2023. PMID: 36806742 Free PMC article.
-
Multi-omics profiling of mouse gastrulation at single-cell resolution.Nature. 2019 Dec;576(7787):487-491. doi: 10.1038/s41586-019-1825-8. Epub 2019 Dec 11. Nature. 2019. PMID: 31827285 Free PMC article.
-
Identification of chromatin states during zebrafish gastrulation using CUT&RUN and CUT&Tag.Dev Dyn. 2022 Apr;251(4):729-742. doi: 10.1002/dvdy.430. Epub 2021 Oct 23. Dev Dyn. 2022. PMID: 34647658 Free PMC article.
-
Redox heterogeneity in mouse embryonic stem cells individualizes cell fate decisions.Dev Cell. 2024 Aug 19;59(16):2118-2133.e8. doi: 10.1016/j.devcel.2024.07.008. Epub 2024 Aug 5. Dev Cell. 2024. PMID: 39106861
-
Rebooting the Epigenomes during Mammalian Early Embryogenesis.Stem Cell Reports. 2020 Dec 8;15(6):1158-1175. doi: 10.1016/j.stemcr.2020.09.005. Epub 2020 Oct 8. Stem Cell Reports. 2020. PMID: 33035464 Free PMC article. Review.
References
-
- Rossant J & Tam PP Emerging asymmetry and embryonic patterning in early mouse development. Dev. Cell 7, 155–164 (2004). - PubMed
-
- Zernicka-Goetz M, Morris SA & Bruce AW Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat. Rev. Genet. 10, 467–477 (2009). - PubMed
-
- Arnold SJ & Robertson EJ Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 10, 91–103 (2009). - PubMed
-
- Lawson KA, Meneses JJ & Pedersen RA Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development 113, 891–911 (1991). - PubMed
-
- Bielinska M, Narita N & Wilson DB Distinct roles for visceral endoderm during embryonic mouse development. Int. J. Dev. Biol. 43, 183–205 (1999). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

