Integrated Cross-Species Analysis Identifies a Conserved Transitional Dendritic Cell Population
- PMID: 31825848
- PMCID: PMC6951814
- DOI: 10.1016/j.celrep.2019.11.042
Integrated Cross-Species Analysis Identifies a Conserved Transitional Dendritic Cell Population
Abstract
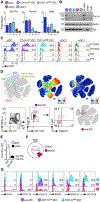
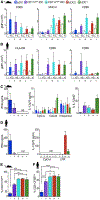
Plasmacytoid dendritic cells (pDCs) are sensor cells with diverse immune functions, from type I interferon (IFN-I) production to antigen presentation, T cell activation, and tolerance. Regulation of these functions remains poorly understood but could be mediated by functionally specialized pDC subpopulations. We address pDC diversity using a high-dimensional single-cell approach: mass cytometry (CyTOF). Our analysis uncovers a murine pDC-like population that specializes in antigen presentation with limited capacity for IFN-I production. Using a multifaceted cross-species comparison, we show that this pDC-like population is the definitive murine equivalent of the recently described human AXL+ DCs, which we unify under the name transitional DCs (tDCs) given their continuum of pDC and cDC2 characteristics. tDCs share developmental traits with pDCs, as well as recruitment dynamics during viral infection. Altogether, we provide a framework for deciphering the function of pDCs and tDCs during diseases, which has the potential to open new avenues for therapeutic design.
Keywords: ASDC; AXL(+) dendritic cells; CyTOF; human; mouse; noncanonical dendritic cells; plasmacytoid dendritic cells; pre-DCs; transitional dendritic cells; viral infection.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures





Similar articles
-
The versatile plasmacytoid dendritic cell: Function, heterogeneity, and plasticity.Int Rev Cell Mol Biol. 2019;349:177-211. doi: 10.1016/bs.ircmb.2019.10.002. Epub 2019 Oct 29. Int Rev Cell Mol Biol. 2019. PMID: 31759431 Review.
-
Mouse strain differences in plasmacytoid dendritic cell frequency and function revealed by a novel monoclonal antibody.J Immunol. 2003 Dec 15;171(12):6466-77. doi: 10.4049/jimmunol.171.12.6466. J Immunol. 2003. PMID: 14662846
-
Phthalates suppress type I interferon in human plasmacytoid dendritic cells via epigenetic regulation.Allergy. 2013 Jul;68(7):870-9. doi: 10.1111/all.12162. Epub 2013 Jun 5. Allergy. 2013. PMID: 23738920
-
Distinctive downmodulation of plasmacytoid dendritic cell functions by vitamin D3 analogue calcipotriol.J Dermatol Sci. 2016 Oct;84(1):71-79. doi: 10.1016/j.jdermsci.2016.06.003. Epub 2016 Jun 3. J Dermatol Sci. 2016. PMID: 27342039
-
Antigen-presentation properties of plasmacytoid dendritic cells.Immunity. 2008 Sep 19;29(3):352-61. doi: 10.1016/j.immuni.2008.09.002. Immunity. 2008. PMID: 18799143 Review.
Cited by
-
The lifespan and kinetics of human dendritic cell subsets and their precursors in health and inflammation.J Exp Med. 2024 Nov 4;221(11):e20220867. doi: 10.1084/jem.20220867. Epub 2024 Oct 17. J Exp Med. 2024. PMID: 39417994
-
Distinct immune cell dynamics correlate with the immunogenicity and reactogenicity of SARS-CoV-2 mRNA vaccine.Cell Rep Med. 2022 May 17;3(5):100631. doi: 10.1016/j.xcrm.2022.100631. Epub 2022 Apr 22. Cell Rep Med. 2022. PMID: 35545084 Free PMC article.
-
Transcriptional regulation of dendritic cell development and function.Front Immunol. 2023 Jul 14;14:1182553. doi: 10.3389/fimmu.2023.1182553. eCollection 2023. Front Immunol. 2023. PMID: 37520521 Free PMC article. Review.
-
The RNase MCPIP3 promotes skin inflammation by orchestrating myeloid cytokine response.Nat Commun. 2021 Jul 2;12(1):4105. doi: 10.1038/s41467-021-24352-w. Nat Commun. 2021. PMID: 34215755 Free PMC article.
-
Skin dendritic cells in melanoma are key for successful checkpoint blockade therapy.J Immunother Cancer. 2021 Jan;9(1):e000832. doi: 10.1136/jitc-2020-000832. J Immunother Cancer. 2021. PMID: 33408092 Free PMC article.
References
-
- Alculumbre SG, Saint-André V, Di Domizio J, Vargas P, Sirven P, Bost P, Maurin M, Maiuri P, Wery M, Roman MS, et al. (2018). Diversification of human plasmacytoid predendritic cells in response to a single stimulus. Nat. Immunol 19, 63–75. - PubMed
-
- Alculumbre S, Raieli S, Hoffmann C, Chelbi R, Danlos F-X, and Soumelis V (2019). Plasmacytoid pre-dendritic cells (pDC): from molecular pathways to function and disease association. Semin. Cell Dev. Biol 86, 24–35. - PubMed
-
- Asselin-Paturel C, Boonstra A, Dalod M, Durand I, Yessaad N, Dezutter-Dambuyant C, Vicari A, O’Garra A, Biron C, Brière F, and Trinchieri G (2001). Mouse type I IFN-producing cells are immature APCs with plasmacytoid morphology. Nat. Immunol 2, 1144–1150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

