Application of metagenomic next-generation sequencing for bronchoalveolar lavage diagnostics in critically ill patients
- PMID: 31813078
- PMCID: PMC7102353
- DOI: 10.1007/s10096-019-03734-5
Application of metagenomic next-generation sequencing for bronchoalveolar lavage diagnostics in critically ill patients
Abstract
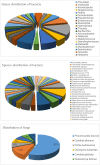
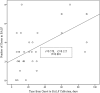
The purpose of this study was to assess the value of metagenomic next-generation sequencing (mNGS) of bronchoalveolar lavage fluid (BALF) for the diagnosis of severe respiratory diseases based on interpretation of sequencing results. BALF samples were harvested and used for mNGS as well as microbiological detection. Infectious bacteria or fungi were defined according to relative abundance and number of unique reads. We performed mNGS on 35 BALF samples from 32 patients. The positive rate reached 100% in the mNGS analysis of nine immunocompromised patients. Compared with the culture method, mNGS had a diagnostic sensitivity of 88.89% and a specificity of 74.07% with an agreement rate of 77.78% between these two methods. Compared with the smear method and PCR, mNGS had a diagnostic sensitivity of 77.78% and a specificity of 70.00%. In 13 cases, detection results were positive by mNGS but negative by culture/smear and PCR. The mNGS findings in 11/32 (34.4%) cases led to changes in treatment strategies. Linear regression analysis showed that diversity was significantly correlated with interval between disease onset and sampling. Dynamic changes in reads could indirectly reflect therapeutic effectiveness. BALF mNGS improves sensitivity of pathogen detection and provides guidance in clinical practice. Potential pathogens can be identified based on relative abundance and number of unique reads.
Keywords: Bronchoalveolar lavage fluid (BALF); Clinical diagnosis; Metagenomic next-generation sequencing (mNGS); Respiratory failure.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
The clinical significance of simultaneous detection of pathogens from bronchoalveolar lavage fluid and blood samples by metagenomic next-generation sequencing in patients with severe pneumonia.J Med Microbiol. 2021 Jan;70(1). doi: 10.1099/jmm.0.001259. J Med Microbiol. 2021. PMID: 33231537
-
The Application of Metagenomic Next-Generation Sequencing in Detection of Pathogen in Bronchoalveolar Lavage Fluid and Sputum Samples of Patients with Pulmonary Infection.Comput Math Methods Med. 2021 Nov 8;2021:7238495. doi: 10.1155/2021/7238495. eCollection 2021. Comput Math Methods Med. 2021. PMID: 34790254 Free PMC article.
-
The diagnostic value of bronchoalveolar lavage fluid metagenomic next-generation sequencing in critically ill patients with respiratory tract infections.Microbiol Spectr. 2024 Aug 6;12(8):e0045824. doi: 10.1128/spectrum.00458-24. Epub 2024 Jun 25. Microbiol Spectr. 2024. PMID: 38916357 Free PMC article.
-
Metagenomic next generation sequencing of bronchoalveolar lavage fluids for the identification of pathogens in patients with pulmonary infection: A retrospective study.Diagn Microbiol Infect Dis. 2024 Sep;110(1):116402. doi: 10.1016/j.diagmicrobio.2024.116402. Epub 2024 Jun 12. Diagn Microbiol Infect Dis. 2024. PMID: 38878340 Review.
-
Diagnostic performance of metagenomic next-generation sequencing for the detection of pathogens in bronchoalveolar lavage fluid in patients with pulmonary infections: Systematic review and meta-analysis.Int J Infect Dis. 2022 Sep;122:867-873. doi: 10.1016/j.ijid.2022.07.054. Epub 2022 Jul 28. Int J Infect Dis. 2022. PMID: 35907477 Review.
Cited by
-
Diagnostic performance of metagenomic sequencing in patients with suspected infection: a large-scale retrospective study.Front Cell Infect Microbiol. 2024 Sep 6;14:1463081. doi: 10.3389/fcimb.2024.1463081. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39310785 Free PMC article.
-
Application of metagenomic next-generation sequencing in the etiological diagnosis of refractory pneumonia in children.Front Microbiol. 2024 Jul 15;15:1357372. doi: 10.3389/fmicb.2024.1357372. eCollection 2024. Front Microbiol. 2024. PMID: 39077741 Free PMC article.
-
Nasal and cutaneous mucormycosis in two patients with lymphoma after chemotherapy and target therapy: Early detection by metagenomic next-generation sequencing.Front Cell Infect Microbiol. 2022 Sep 16;12:960766. doi: 10.3389/fcimb.2022.960766. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36189372 Free PMC article.
-
Severe pediatric Mycoplasma pneumonia as the cause of diffuse alveolar hemorrhage requiring veno-venous extracorporeal membrane oxygenation: A case report.Front Pediatr. 2023 Jan 6;10:925655. doi: 10.3389/fped.2022.925655. eCollection 2022. Front Pediatr. 2023. PMID: 36683817 Free PMC article.
-
Clinical Characteristics of Chronic Lung Abscess Associated with Parvimonas micra Diagnosed Using Metagenomic Next-Generation Sequencing.Infect Drug Resist. 2021 Mar 23;14:1191-1198. doi: 10.2147/IDR.S304569. eCollection 2021. Infect Drug Resist. 2021. PMID: 33790589 Free PMC article.
References
-
- Leo Stefano, Gaïa Nadia, Ruppé Etienne, Emonet Stephane, Girard Myriam, Lazarevic Vladimir, Schrenzel Jacques. Detection of Bacterial Pathogens from Broncho-Alveolar Lavage by Next-Generation Sequencing. International Journal of Molecular Sciences. 2017;18(9):2011. doi: 10.3390/ijms18092011. - DOI - PMC - PubMed
-
- Meyer KC, Raghu G, Baughman RP, Brown KK, Costabel U, du Bois RM, et al. An official American Thoracic Society clinical practice guideline: the clinical utility of bronchoalveolar lavage cellular analysis in interstitial lung disease. Am J Respir Crit Care Med. 2012;185:1004–1014. doi: 10.1164/rccm.201202-0320ST. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

