Structural basis for shieldin complex subunit 3-mediated recruitment of the checkpoint protein REV7 during DNA double-strand break repair
- PMID: 31796627
- PMCID: PMC6952594
- DOI: 10.1074/jbc.RA119.011464
Structural basis for shieldin complex subunit 3-mediated recruitment of the checkpoint protein REV7 during DNA double-strand break repair
Abstract
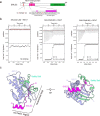
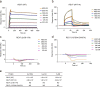
Shieldin complex subunit 3 (SHLD3) is the apical subunit of a recently-identified shieldin complex and plays a critical role in DNA double-strand break repair. To fulfill its function in DNA repair, SHLD3 interacts with the mitotic spindle assembly checkpoint protein REV7 homolog (REV7), but the details of this interaction remain obscure. Here, we present the crystal structures of REV7 in complex with SHLD3's REV7-binding domain (RBD) at 2.2-2.3 Å resolutions. The structures revealed that the ladle-shaped RBD in SHLD3 uses its N-terminal loop and C-terminal α-helix (αC-helix) in its interaction with REV7. The N-terminal loop exhibited a structure similar to those previously identified in other REV7-binding proteins, and the less-conserved αC-helix region adopted a distinct mode for binding REV7. In vitro and in vivo binding analyses revealed that the N-terminal loop and the αC-helix are both indispensable for high-affinity REV7 binding (with low-nanomolar affinity), underscoring the crucial role of SHLD3 αC-helix in protein binding. Moreover, binding kinetics analyses revealed that the REV7 "safety belt" region, which plays a role in binding other proteins, is essential for SHLD3-REV7 binding, as this region retards the dissociation of the RBD from the bound REV7. Together, the findings of our study reveal the molecular basis of the SHLD3-REV7 interaction and provide critical insights into how SHLD3 recognizes REV7.
Keywords: DNA repair; REV7; crystal structure; isothermal titration calorimetry (ITC); mitotic arrest deficient 2 like 2 (MAD2L2); nonhomologous end-joining repair; protein–protein interaction; shieldin complex subunit 3 (SHLD3); structural biology; surface plasmon resonance (SPR).
© 2020 Dai et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


Similar articles
-
Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex.Proc Natl Acad Sci U S A. 2021 Feb 23;118(8):e2024512118. doi: 10.1073/pnas.2024512118. Proc Natl Acad Sci U S A. 2021. PMID: 33597306 Free PMC article.
-
Molecular basis for assembly of the shieldin complex and its implications for NHEJ.Nat Commun. 2020 Apr 24;11(1):1972. doi: 10.1038/s41467-020-15879-5. Nat Commun. 2020. PMID: 32332881 Free PMC article.
-
Shieldin complex assembly kinetics and DNA binding by SHLD3.Commun Biol. 2023 Apr 8;6(1):384. doi: 10.1038/s42003-023-04757-7. Commun Biol. 2023. PMID: 37031298 Free PMC article.
-
REV7: Jack of many trades.Trends Cell Biol. 2021 Aug;31(8):686-701. doi: 10.1016/j.tcb.2021.04.002. Epub 2021 May 4. Trends Cell Biol. 2021. PMID: 33962851 Free PMC article. Review.
-
REV7 directs DNA repair pathway choice.Trends Cell Biol. 2021 Dec;31(12):965-978. doi: 10.1016/j.tcb.2021.05.009. Epub 2021 Jun 16. Trends Cell Biol. 2021. PMID: 34147298 Free PMC article. Review.
Cited by
-
Genetic Variants in Double-Strand Break Repair Pathway Genes to Predict Platinum-Based Chemotherapy Prognosis in Patients With Lung Cancer.Front Pharmacol. 2022 Jul 11;13:915822. doi: 10.3389/fphar.2022.915822. eCollection 2022. Front Pharmacol. 2022. PMID: 35899106 Free PMC article.
-
The dystonia gene THAP1 controls DNA double-strand break repair choice.Mol Cell. 2021 Jun 17;81(12):2611-2624.e10. doi: 10.1016/j.molcel.2021.03.034. Epub 2021 Apr 14. Mol Cell. 2021. PMID: 33857404 Free PMC article.
-
MAD2L2 dimerization and TRIP13 control shieldin activity in DNA repair.Nat Commun. 2021 Sep 14;12(1):5421. doi: 10.1038/s41467-021-25724-y. Nat Commun. 2021. PMID: 34521823 Free PMC article.
-
p31comet promotes homologous recombination by inactivating REV7 through the TRIP13 ATPase.Proc Natl Acad Sci U S A. 2020 Oct 27;117(43):26795-26803. doi: 10.1073/pnas.2008830117. Epub 2020 Oct 13. Proc Natl Acad Sci U S A. 2020. PMID: 33051298 Free PMC article.
-
Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex.Proc Natl Acad Sci U S A. 2021 Feb 23;118(8):e2024512118. doi: 10.1073/pnas.2024512118. Proc Natl Acad Sci U S A. 2021. PMID: 33597306 Free PMC article.
References
-
- Bunting S. F., Callén E., Wong N., Chen H.-T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., Xu X., Deng C.-X., Finkel T., Nussenzweig M., Stark J. M., and Nussenzweig A. (2010) 53BP1 Inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 10.1016/j.cell.2010.03.012 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

