Metabolic Incorporation of N-Acetyl Muramic Acid Probes into Bacterial Peptidoglycan
- PMID: 31763799
- PMCID: PMC7591266
- DOI: 10.1002/cpch.74
Metabolic Incorporation of N-Acetyl Muramic Acid Probes into Bacterial Peptidoglycan
Abstract
Bacterial cells utilize small carbohydrate building blocks to construct peptidoglycan (PG), a highly conserved mesh-like polymer that serves as a protective coat for the cell. PG production has long been a target for antibiotics, and its breakdown is a source for human immune recognition. A key component of bacterial PG, N-acetyl muramic acid (NAM), is a vital element in many synthetically derived immunostimulatory compounds. However, the exact molecular details of these structures and how they are generated remain unknown due to a lack of chemical probes surrounding the NAM core. A robust synthetic strategy to generate bioorthogonally tagged NAM carbohydrate units is implemented. These molecules serve as precursors for PG biosynthesis and recycling. Escherichia coli cells are metabolically engineered to incorporate the bioorthogonal NAM probes into their PG network. The probes are subsequently modified using copper-catalyzed azide-alkyne cycloaddition to install fluorophores directly into the bacterial PG, as confirmed by super-resolution microscopy and high-resolution mass spectrometry. Here, synthetic notes for key elements of this process to generate the sugar probes as well as streamlined user-friendly metabolic labeling strategies for both microbiology and immunological applications are described. © 2019 by John Wiley & Sons, Inc. Basic Protocol 1: Synthesis of peracetylated 2-azido glucosamine Basic Protocol 2: Synthesis of 2-azido and 2-alkyne NAM Basic Protocol 3: Synthesis of 3-azido NAM methyl ester Basic Protocol 4: Incorporation of NAM probes into bacterial peptidoglycan Basic Protocol 5: Confirmation of bacterial cell wall remodeling by mass spectrometry.
Keywords: bacterial peptidoglycan; bioorthogonal chemistry; carbohydrates; click chemistry; fluorescent labeling; mass spectrometry; metabolic incorporation; microscopy.
© 2019 John Wiley & Sons, Inc.
Figures
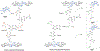


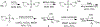
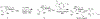
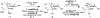


Similar articles
-
Protected N-Acetyl Muramic Acid Probes Improve Bacterial Peptidoglycan Incorporation via Metabolic Labeling.ACS Chem Biol. 2021 Oct 15;16(10):1908-1916. doi: 10.1021/acschembio.1c00268. Epub 2021 Sep 10. ACS Chem Biol. 2021. PMID: 34506714 Free PMC article.
-
Metabolic labelling of the carbohydrate core in bacterial peptidoglycan and its applications.Nat Commun. 2017 Apr 20;8:15015. doi: 10.1038/ncomms15015. Nat Commun. 2017. PMID: 28425464 Free PMC article.
-
Minimalist Tetrazine N-Acetyl Muramic Acid Probes for Rapid and Efficient Labeling of Commensal and Pathogenic Peptidoglycans in Living Bacterial Culture and During Macrophage Invasion.J Am Chem Soc. 2024 Mar 13;146(10):6817-6829. doi: 10.1021/jacs.3c13644. Epub 2024 Mar 1. J Am Chem Soc. 2024. PMID: 38427023 Free PMC article.
-
d-Amino Acid Derivatives as in Situ Probes for Visualizing Bacterial Peptidoglycan Biosynthesis.Acc Chem Res. 2019 Sep 17;52(9):2713-2722. doi: 10.1021/acs.accounts.9b00311. Epub 2019 Aug 16. Acc Chem Res. 2019. PMID: 31419110 Review.
-
Recent Advances in the Synthesis and Biological Applications of Peptidoglycan Fragments.Chemistry. 2022 Aug 1;28(43):e202200788. doi: 10.1002/chem.202200788. Epub 2022 Jun 16. Chemistry. 2022. PMID: 35560956 Review.
Cited by
-
Rapid Production of Multifunctional Self-Assembling Peptides for Incorporation and Visualization within Hydrogel Biomaterials.ACS Biomater Sci Eng. 2021 Sep 13;7(9):4175-4195. doi: 10.1021/acsbiomaterials.1c00589. Epub 2021 Jul 20. ACS Biomater Sci Eng. 2021. PMID: 34283566 Free PMC article.
-
Imitate to illuminate: labeling of bacterial peptidoglycan with fluorescent and bio-orthogonal stem peptide-mimicking probes.RSC Chem Biol. 2022 Aug 5;3(10):1198-1208. doi: 10.1039/d2cb00086e. eCollection 2022 Oct 5. RSC Chem Biol. 2022. PMID: 36320889 Free PMC article. Review.
-
Inhibition of Streptococcus pneumoniae growth by masarimycin.Microbiology (Reading). 2022 Apr;168(4):001182. doi: 10.1099/mic.0.001182. Microbiology (Reading). 2022. PMID: 35467499 Free PMC article.
-
Protected N-Acetyl Muramic Acid Probes Improve Bacterial Peptidoglycan Incorporation via Metabolic Labeling.ACS Chem Biol. 2021 Oct 15;16(10):1908-1916. doi: 10.1021/acschembio.1c00268. Epub 2021 Sep 10. ACS Chem Biol. 2021. PMID: 34506714 Free PMC article.
-
Localizing Peptidoglycan Synthesis in Helicobacter pylori using Clickable Metabolic Probes.Curr Protoc. 2021 Apr;1(4):e80. doi: 10.1002/cpz1.80. Curr Protoc. 2021. PMID: 33844460 Free PMC article.
References
-
- Barreteau H, Kovač A, Boniface A, Sova M, Gobec S, and Blanot D. 2008. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiology Reviews. - PubMed
-
- Bouhss A, Crouvoisier M, Blanot D, and Mengin-Lecreulx D. 2004. Purification and characterization of the bacterial MraY translocase catalyzing the first membrane step of peptidoglycan biosynthesis. Journal of Biological Chemistry. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

