Structural basis of homologous recombination
- PMID: 31748913
- PMCID: PMC6957567
- DOI: 10.1007/s00018-019-03365-1
Structural basis of homologous recombination
Abstract
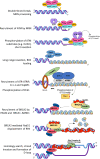
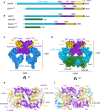
Homologous recombination (HR) is a pathway to faithfully repair DNA double-strand breaks (DSBs). At the core of this pathway is a DNA recombinase, which, as a nucleoprotein filament on ssDNA, pairs with homologous DNA as a template to repair the damaged site. In eukaryotes Rad51 is the recombinase capable of carrying out essential steps including strand invasion, homology search on the sister chromatid and strand exchange. Importantly, a tightly regulated process involving many protein factors has evolved to ensure proper localisation of this DNA repair machinery and its correct timing within the cell cycle. Dysregulation of any of the proteins involved can result in unchecked DNA damage, leading to uncontrolled cell division and cancer. Indeed, many are tumour suppressors and are key targets in the development of new cancer therapies. Over the past 40 years, our structural and mechanistic understanding of homologous recombination has steadily increased with notable recent advancements due to the advances in single particle cryo electron microscopy. These have resulted in higher resolution structural models of the signalling proteins ATM (ataxia telangiectasia mutated), and ATR (ataxia telangiectasia and Rad3-related protein), along with various structures of Rad51. However, structural information of the other major players involved, such as BRCA1 (breast cancer type 1 susceptibility protein) and BRCA2 (breast cancer type 2 susceptibility protein), has been limited to crystal structures of isolated domains and low-resolution electron microscopy reconstructions of the full-length proteins. Here we summarise the current structural understanding of homologous recombination, focusing on key proteins in recruitment and signalling events as well as the mediators for the Rad51 recombinase.
Keywords: Cryo electron microscopy; DNA damage signalling and repair; Double-strand break repair; Homologous recombination; X-ray crystallography.
Figures



Similar articles
-
Dissecting the Recombination Mediator Activity of BRCA2 Using Biochemical Methods.Methods Enzymol. 2018;600:479-511. doi: 10.1016/bs.mie.2017.11.018. Methods Enzymol. 2018. PMID: 29458771
-
BRCA1 regulates RAD51 function in response to DNA damage and suppresses spontaneous sister chromatid replication slippage: implications for sister chromatid cohesion, genome stability, and carcinogenesis.Cancer Res. 2005 Dec 15;65(24):11384-91. doi: 10.1158/0008-5472.CAN-05-2156. Cancer Res. 2005. PMID: 16357146
-
The BRCA Tumor Suppressor Network in Chromosome Damage Repair by Homologous Recombination.Annu Rev Biochem. 2019 Jun 20;88:221-245. doi: 10.1146/annurev-biochem-013118-111058. Epub 2019 Mar 27. Annu Rev Biochem. 2019. PMID: 30917004 Free PMC article. Review.
-
The Post-Synaptic Function of Brca2.Sci Rep. 2019 Mar 14;9(1):4554. doi: 10.1038/s41598-019-41054-y. Sci Rep. 2019. PMID: 30872704 Free PMC article.
-
Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins.Cold Spring Harb Perspect Biol. 2015 Apr 1;7(4):a016600. doi: 10.1101/cshperspect.a016600. Cold Spring Harb Perspect Biol. 2015. PMID: 25833843 Free PMC article. Review.
Cited by
-
BRCA Mutations in Prostate Cancer: Assessment, Implications and Treatment Considerations.Int J Mol Sci. 2021 Nov 23;22(23):12628. doi: 10.3390/ijms222312628. Int J Mol Sci. 2021. PMID: 34884434 Free PMC article. Review.
-
In vivo tracking of functionally tagged Rad51 unveils a robust strategy of homology search.Nat Struct Mol Biol. 2023 Oct;30(10):1582-1591. doi: 10.1038/s41594-023-01065-w. Epub 2023 Aug 21. Nat Struct Mol Biol. 2023. PMID: 37605042
-
Olaparib-Resistant BRCA2MUT Ovarian Cancer Cells with Restored BRCA2 Abrogate Olaparib-Induced DNA Damage and G2/M Arrest Controlled by the ATR/CHK1 Pathway for Survival.Cells. 2023 Mar 29;12(7):1038. doi: 10.3390/cells12071038. Cells. 2023. PMID: 37048111 Free PMC article.
-
Targeting Neoantigens in Cancer: Possibilities and Opportunities in Breast Cancer.Antibodies (Basel). 2024 Jun 10;13(2):46. doi: 10.3390/antib13020046. Antibodies (Basel). 2024. PMID: 38920970 Free PMC article. Review.
-
The multifaceted roles of DNA repair and replication proteins in aging and obesity.DNA Repair (Amst). 2021 Mar;99:103049. doi: 10.1016/j.dnarep.2021.103049. Epub 2021 Jan 21. DNA Repair (Amst). 2021. PMID: 33529944 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

