Genome-Wide Transcriptome Analysis of Human Papillomavirus 16-Infected Primary Keratinocytes Reveals Subtle Perturbations Mostly due to E7 Protein Expression
- PMID: 31748387
- PMCID: PMC7000963
- DOI: 10.1128/JVI.01360-19
Genome-Wide Transcriptome Analysis of Human Papillomavirus 16-Infected Primary Keratinocytes Reveals Subtle Perturbations Mostly due to E7 Protein Expression
Erratum in
-
Correction for Bienkowska-Haba et al., "Genome-Wide Transcriptome Analysis of Human Papillomavirus 16-Infected Primary Keratinocytes Reveals Subtle Perturbations Mostly Due To E7 Protein Expression".J Virol. 2022 Nov 23;96(22):e0165922. doi: 10.1128/jvi.01659-22. Epub 2022 Nov 7. J Virol. 2022. PMID: 36342294 Free PMC article. No abstract available.
Abstract
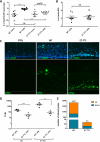
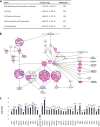
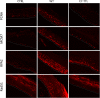
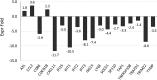
It is established that the host cell transcriptomes of natural lesions, organotypic rafts, and human papillomavirus (HPV)-immortalized keratinocytes are altered in the presence of HPV genomes. However, the establishment of HPV-harboring cell lines requires selection and immortalization, which makes it impossible to distinguish between alterations directly induced by HPV or indirectly by the need for immortalization or selection. To address direct effects of HPV infection on the host cell transcriptome, we have used our recently established infection model that allows efficient infection of primary keratinocytes with HPV16 virions. We observed only a small set of genes to be deregulated at the transcriptional level at 7 days postinfection (dpi), most of which fall into the category regulated by pocket proteins pRb, p107, and p130. Furthermore, cell cycle genes were not deregulated in cells infected with a virus lacking E7 despite the presence of episomal genome and viral transcripts. These findings imply that the majority of transcriptional changes are due to the E7 protein impairing pocket protein function. Additional pathways, such as the Fanconi anemia-BRCA pathway, became perturbed only after long-term culturing of infected cells. When grown as organotypic raft cultures, keratinocytes infected with wild-type but not E7 mutant virus had perturbed transcriptional regulation of pathways previously identified in natural lesions and in rafts derived from immortalized keratinocytes. We conclude that the HPV infection model provides a valuable tool to distinguish immediate transcriptional alterations from those induced by persistent infection and the need for selection and immortalization.IMPORTANCE To establish infection and complete the viral life cycle, human papillomavirus (HPV) needs to alter the transcriptional program of host cells. Until recently, studies were restricted to keratinocyte-derived cell lines immortalized by HPV due to the lack of experimental systems to efficiently infect primary keratinocytes. Need for selection and immortalization made it impossible to distinguish between alterations induced by HPV and secondary adaptation due to selection and immortalization. With our recent establishment of an extracellular matrix (ECM)-to-cell transfer system allowing efficient infection of primary keratinocytes, we were able to identify transcriptional changes attributable to HPV16 infection. Most perturbed genes fall into the class of S-phase genes, which are regulated by pocket proteins. Indeed, infection with viruses lacking E7 abrogated most transcriptional changes. It is important to note that many transcriptional alterations thought to be important for the HPV life cycle are actually late events that may reflect immortalization and, possibly, disease progression.
Keywords: E7; HPV16; RB; infection model; organotypic raft culture; p53; pocket protein; primary foreskin keratinocytes; transcriptome.
Copyright © 2020 American Society for Microbiology.
Figures
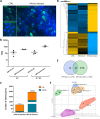
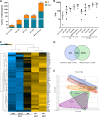
Similar articles
-
Inhibition of Epstein-Barr Virus Replication in Human Papillomavirus-Immortalized Keratinocytes.J Virol. 2019 Jan 4;93(2):e01216-18. doi: 10.1128/JVI.01216-18. Print 2019 Jan 15. J Virol. 2019. PMID: 30381489 Free PMC article.
-
A new cell culture model to genetically dissect the complete human papillomavirus life cycle.PLoS Pathog. 2018 Mar 1;14(3):e1006846. doi: 10.1371/journal.ppat.1006846. eCollection 2018 Mar. PLoS Pathog. 2018. PMID: 29494681 Free PMC article.
-
Epstein-Barr virus replication within differentiated epithelia requires pRb sequestration of activator E2F transcription factors.J Virol. 2024 Oct 22;98(10):e0099524. doi: 10.1128/jvi.00995-24. Epub 2024 Sep 18. J Virol. 2024. PMID: 39291960
-
Activation of telomerase by HPVs.Virus Res. 2017 Mar 2;231:50-55. doi: 10.1016/j.virusres.2016.11.003. Epub 2016 Nov 15. Virus Res. 2017. PMID: 27863966 Review.
-
The human papillomavirus E7 oncoprotein as a regulator of transcription.Virus Res. 2017 Mar 2;231:56-75. doi: 10.1016/j.virusres.2016.10.017. Epub 2016 Nov 8. Virus Res. 2017. PMID: 27818212 Free PMC article. Review.
Cited by
-
Experimental Support for Human Papillomavirus Genome Amplification Early after Infectious Delivery.J Virol. 2023 Jun 29;97(6):e0021423. doi: 10.1128/jvi.00214-23. Epub 2023 May 18. J Virol. 2023. PMID: 37223953 Free PMC article.
-
Human Papillomavirus Genome Copy Number Is Maintained by S-Phase Amplification, Genome Loss to the Cytosol during Mitosis, and Degradation in G1 Phase.J Virol. 2023 Feb 28;97(2):e0187922. doi: 10.1128/jvi.01879-22. Epub 2023 Feb 7. J Virol. 2023. PMID: 36749071 Free PMC article.
-
Conditional Cell Reprogramming and Air-Liquid Interface Modeling Life Cycle of Oncogenic Viruses (HPV and EBV) in Epithelial Cells and Virus-Associated Human Carcinomas.Viruses. 2023 Jun 17;15(6):1388. doi: 10.3390/v15061388. Viruses. 2023. PMID: 37376685 Free PMC article. Review.
-
Retinoblastoma Protein Is Required for Epstein-Barr Virus Replication in Differentiated Epithelia.J Virol. 2023 Feb 28;97(2):e0103222. doi: 10.1128/jvi.01032-22. Epub 2023 Jan 31. J Virol. 2023. PMID: 36719239 Free PMC article.
-
HPV induced R-loop formation represses innate immune gene expression while activating DNA damage repair pathways.PLoS Pathog. 2024 Aug 23;20(8):e1012454. doi: 10.1371/journal.ppat.1012454. eCollection 2024 Aug. PLoS Pathog. 2024. PMID: 39178326 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

