AZD7648 is a potent and selective DNA-PK inhibitor that enhances radiation, chemotherapy and olaparib activity
- PMID: 31699977
- PMCID: PMC6838110
- DOI: 10.1038/s41467-019-12836-9
AZD7648 is a potent and selective DNA-PK inhibitor that enhances radiation, chemotherapy and olaparib activity
Abstract
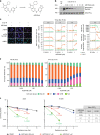
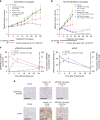
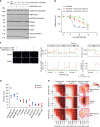
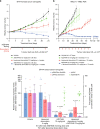
DNA-dependent protein kinase (DNA-PK) is a critical player in the DNA damage response (DDR) and instrumental in the non-homologous end-joining pathway (NHEJ) used to detect and repair DNA double-strand breaks (DSBs). We demonstrate that the potent and highly selective DNA-PK inhibitor, AZD7648, is an efficient sensitizer of radiation- and doxorubicin-induced DNA damage, with combinations in xenograft and patient-derived xenograft (PDX) models inducing sustained regressions. Using ATM-deficient cells, we demonstrate that AZD7648, in combination with the PARP inhibitor olaparib, increases genomic instability, resulting in cell growth inhibition and apoptosis. AZD7648 enhanced olaparib efficacy across a range of doses and schedules in xenograft and PDX models, enabling sustained tumour regression and providing a clear rationale for its clinical investigation. Through its differentiated mechanism of action as an NHEJ inhibitor, AZD7648 complements the current armamentarium of DDR-targeted agents and has potential in combination with these agents to achieve deeper responses to current therapies.
Conflict of interest statement
J.H.L.F., A.R.-M., M.V.-C., P.W.G.W., V.F., N.J., P.M.F., A.K., S.E.W., J.C., J.N., D.B., G.M.L., M.R.V.F., J.W., A.S., L.O.O.C., S.L., S.E.F., M.J.O.C., S.J.H., E.D., F.W.G., B.R.D. and E.B.C. are or were employees of AstraZeneca at the time of conducting these studies. All authors hold stock in AstraZeneca.
Figures
Similar articles
-
DNA-PK inhibitor AZD7648 is a more portent radiosensitizer than PARP inhibitor Olaparib in BRCA1/2 deficient tumors.DNA Repair (Amst). 2024 Jul;139:103689. doi: 10.1016/j.dnarep.2024.103689. Epub 2024 May 6. DNA Repair (Amst). 2024. PMID: 38749239
-
The DNA-PK Inhibitor AZD7648 Sensitizes Patient-Derived Ovarian Cancer Xenografts to Pegylated Liposomal Doxorubicin and Olaparib Preventing Abdominal Metastases.Mol Cancer Ther. 2022 Apr 1;21(4):555-567. doi: 10.1158/1535-7163.MCT-21-0420. Mol Cancer Ther. 2022. PMID: 35149547
-
Inhibition of DNA-PK with AZD7648 Sensitizes Tumor Cells to Radiotherapy and Induces Type I IFN-Dependent Durable Tumor Control.Clin Cancer Res. 2021 Aug 1;27(15):4353-4366. doi: 10.1158/1078-0432.CCR-20-3701. Epub 2021 May 19. Clin Cancer Res. 2021. PMID: 34011558 Free PMC article.
-
Development and therapeutic potential of DNA-dependent protein kinase inhibitors.Bioorg Chem. 2024 Sep;150:107608. doi: 10.1016/j.bioorg.2024.107608. Epub 2024 Jun 29. Bioorg Chem. 2024. PMID: 38981210 Review.
-
Clinically Applicable Inhibitors Impacting Genome Stability.Molecules. 2018 May 13;23(5):1166. doi: 10.3390/molecules23051166. Molecules. 2018. PMID: 29757235 Free PMC article. Review.
Cited by
-
Genomic, Transcriptomic, and Functional Alterations in DNA Damage Response Pathways as Putative Biomarkers of Chemotherapy Response in Ovarian Cancer.Cancers (Basel). 2021 Mar 20;13(6):1420. doi: 10.3390/cancers13061420. Cancers (Basel). 2021. PMID: 33804647 Free PMC article.
-
Disparate pathways for extrachromosomal DNA biogenesis and genomic DNA repair.bioRxiv [Preprint]. 2023 Oct 23:2023.10.22.563489. doi: 10.1101/2023.10.22.563489. bioRxiv. 2023. Update in: Cancer Discov. 2025 Jan 13;15(1):69-82. doi: 10.1158/2159-8290.CD-23-1117 PMID: 37961138 Free PMC article. Updated. Preprint.
-
Mismatch repair-proficient tumor footprints in the sands of immune desert: mechanistic constraints and precision platforms.Front Immunol. 2024 Jul 19;15:1414376. doi: 10.3389/fimmu.2024.1414376. eCollection 2024. Front Immunol. 2024. PMID: 39100682 Free PMC article. Review.
-
Development and Evolution of DNA-Dependent Protein Kinase Inhibitors toward Cancer Therapy.Int J Mol Sci. 2022 Apr 12;23(8):4264. doi: 10.3390/ijms23084264. Int J Mol Sci. 2022. PMID: 35457081 Free PMC article. Review.
-
Polλ promotes microhomology-mediated end-joining.Nat Struct Mol Biol. 2023 Jan;30(1):107-114. doi: 10.1038/s41594-022-00895-4. Epub 2022 Dec 19. Nat Struct Mol Biol. 2023. PMID: 36536104 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

