Meta-Analysis of Gene Expression Changes in the Blood of Patients with Mild Cognitive Impairment and Alzheimer's Disease Dementia
- PMID: 31671574
- PMCID: PMC6862214
- DOI: 10.3390/ijms20215403
Meta-Analysis of Gene Expression Changes in the Blood of Patients with Mild Cognitive Impairment and Alzheimer's Disease Dementia
Abstract
Background: Dementia is a major public health concern affecting approximately 47 million people worldwide. Mild cognitive impairment (MCI) is one form of dementia that affects an individual's memory with or without affecting their daily life. Alzheimer's disease dementia (ADD) is a more severe form of dementia that usually affects elderly individuals. It remains unclear whether MCI is a distinct disorder from or an early stage of ADD.
Methods: Gene expression data from blood were analyzed to identify potential biomarkers that may be useful for distinguishing between these two forms of dementia.
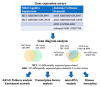
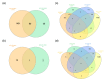
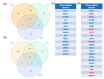
Results: A meta-analysis revealed 91 genes dysregulated in individuals with MCI and 387 genes dysregulated in ADD. Pathway analysis identified seven pathways shared between MCI and ADD and nine ADD-specific pathways. Fifteen transcription factors were associated with MCI and ADD, whereas seven transcription factors were specific for ADD. Mir-335-5p was specific for ADD, suggesting that it may be useful as a biomarker. Diseases that are associated with MCI and ADD included developmental delays, cognition impairment, and movement disorders.
Conclusion: These results provide a better molecular understanding of peripheral changes that occur in MCI and ADD patients and may be useful in the identification of diagnostic and prognostic biomarkers.
Keywords: Alzheimer’s disease; dementia; gene expression; mild cognitive impairment; network analysis.
Conflict of interest statement
The authors have no conflict of interest to report. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

Similar articles
-
MicroRNA Expression Signature in Mild Cognitive Impairment Due to Alzheimer's Disease.Mol Neurobiol. 2020 Nov;57(11):4408-4416. doi: 10.1007/s12035-020-02029-7. Epub 2020 Jul 31. Mol Neurobiol. 2020. PMID: 32737762 Free PMC article.
-
Altered microRNAs related to synaptic function as potential plasma biomarkers for Alzheimer's disease.Alzheimers Res Ther. 2019 May 15;11(1):46. doi: 10.1186/s13195-019-0501-4. Alzheimers Res Ther. 2019. PMID: 31092279 Free PMC article.
-
Increased prediction value of biomarker combinations for the conversion of mild cognitive impairment to Alzheimer's dementia.Transl Neurodegener. 2020 Aug 3;9(1):30. doi: 10.1186/s40035-020-00210-5. Transl Neurodegener. 2020. PMID: 32741361 Free PMC article.
-
Circulating microRNAs as Biomarkers of Alzheimer's Disease: A Systematic Review.J Alzheimers Dis. 2016;49(3):755-66. doi: 10.3233/JAD-150619. J Alzheimers Dis. 2016. PMID: 26484928 Review.
-
Evaluation of the diagnostic value of peripheral BDNF levels for Alzheimer's disease and mild cognitive impairment: results of a meta-analysis.Int J Neurosci. 2020 Mar;130(3):218-230. doi: 10.1080/00207454.2019.1667794. Epub 2019 Sep 24. Int J Neurosci. 2020. PMID: 31518516 Review.
Cited by
-
Cytosolic calcium: Judge, jury and executioner of neurodegeneration in Alzheimer's disease and beyond.Alzheimers Dement. 2023 Aug;19(8):3701-3717. doi: 10.1002/alz.13065. Epub 2023 May 3. Alzheimers Dement. 2023. PMID: 37132525 Free PMC article. Review.
-
A Comparison of Gene Expression Changes in the Blood of Individuals Consuming Diets Supplemented with Olives, Nuts or Long-Chain Omega-3 Fatty Acids.Nutrients. 2020 Dec 8;12(12):3765. doi: 10.3390/nu12123765. Nutrients. 2020. PMID: 33302351 Free PMC article.
-
A Comparative Cross-Platform Analysis to Identify Potential Biomarker Genes for Evaluation of Teratozoospermia and Azoospermia.Genes (Basel). 2022 Sep 25;13(10):1721. doi: 10.3390/genes13101721. Genes (Basel). 2022. PMID: 36292606 Free PMC article.
-
Key Genes and Biochemical Networks in Various Brain Regions Affected in Alzheimer's Disease.Cells. 2022 Mar 14;11(6):987. doi: 10.3390/cells11060987. Cells. 2022. PMID: 35326437 Free PMC article.
-
Bioinformatic Analysis Reveals Phosphodiesterase 4D-Interacting Protein as a Key Frontal Cortex Dementia Switch Gene.Int J Mol Sci. 2020 May 27;21(11):3787. doi: 10.3390/ijms21113787. Int J Mol Sci. 2020. PMID: 32471155 Free PMC article.
References
-
- Jack C.R., Jr., Bennett D.A., Blennow K., Carrillo M.C., Dunn B., Haeberlein S.B., Holtzman D.M., Jagust W., Jessen F., Karlawish J., et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14:535–562. doi: 10.1016/j.jalz.2018.02.018. - DOI - PMC - PubMed
-
- Jack C.R., Jr., Therneau T.M., Weigand S.D., Wiste H.J., Knopman D.S., Vemuri P., Lowe V.J., Mielke M.M., Roberts R.O., Machulda M.M., et al. Prevalence of Biologically vs Clinically Defined Alzheimer Spectrum Entities Using the National Institute on Aging-Alzheimer’s Association Research Framework. JAMA Neurol. 2019;76:1174–1183. doi: 10.1001/jamaneurol.2019.1971. - DOI - PMC - PubMed
-
- Bennett D.A., Wilson R.S., Schneider J.A., Evans D.A., Aggarwal N.T., Arnold S.E., Cochran E.J., Berry-Kravis E., Bienias J.L. Apolipoprotein E epsilon4 allele, AD pathology, and the clinical expression of Alzheimer’s disease. Neurology. 2003;60:246–252. doi: 10.1212/01.WNL.0000042478.08543.F7. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

