Identification of a monoclonal antibody that targets PD-1 in a manner requiring PD-1 Asn58 glycosylation
- PMID: 31667366
- PMCID: PMC6814707
- DOI: 10.1038/s42003-019-0642-9
Identification of a monoclonal antibody that targets PD-1 in a manner requiring PD-1 Asn58 glycosylation
Abstract
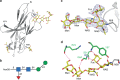
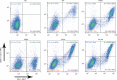
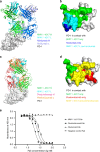
Programmed cell death 1 (PD-1) is inhibitory receptor and immune checkpoint protein. Blocking the interaction of PD-1 and its ligands PD-L1/ L2 is able to active T-cell-mediated antitumor response. Monoclonal antibody-based drugs targeting PD-1 pathway have exhibited great promise in cancer therapy. Here we show that MW11-h317, an anti-PD-1 monoclonal antibody, displays high affinity for PD-1 and blocks PD-1 interactions with PD-L1/L2. MW11-h317 can effectively induce T-cell-mediated immune response and inhibit tumor growth in mouse model. Crystal structure of PD-1/MW11-h317 Fab complex reveals that both the loops and glycosylation of PD-1 are involved in recognition and binding, in which Asn58 glycosylation plays a critical role. The unique glycan epitope in PD-1 to MW11-h317 is different from the first two approved clinical PD-1 antibodies, nivolumab and pembrolizumab. These results suggest MW11-h317 as a therapeutic monoclonal antibody of PD-1 glycosylation-targeting which may become efficient alternative for cancer therapy.
Keywords: Cancer; Cancer immunotherapy; Structural biology.
© The Author(s) 2019.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures
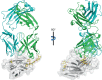


Similar articles
-
An unexpected N-terminal loop in PD-1 dominates binding by nivolumab.Nat Commun. 2017 Feb 6;8:14369. doi: 10.1038/ncomms14369. Nat Commun. 2017. PMID: 28165004 Free PMC article.
-
Biophysical and Immunological Characterization and In Vivo Pharmacokinetics and Toxicology in Nonhuman Primates of the Anti-PD-1 Antibody Pembrolizumab.Mol Cancer Ther. 2020 Jun;19(6):1298-1307. doi: 10.1158/1535-7163.MCT-19-0774. Epub 2020 Mar 30. Mol Cancer Ther. 2020. PMID: 32229606
-
Structural Biology of the Immune Checkpoint Receptor PD-1 and Its Ligands PD-L1/PD-L2.Structure. 2017 Aug 1;25(8):1163-1174. doi: 10.1016/j.str.2017.06.011. Structure. 2017. PMID: 28768162 Review.
-
Structural basis of HLX10 PD-1 receptor recognition, a promising anti-PD-1 antibody clinical candidate for cancer immunotherapy.PLoS One. 2021 Dec 31;16(12):e0257972. doi: 10.1371/journal.pone.0257972. eCollection 2021. PLoS One. 2021. PMID: 34972111 Free PMC article.
-
Molecular Interactions of Antibody Drugs Targeting PD-1, PD-L1, and CTLA-4 in Immuno-Oncology.Molecules. 2019 Mar 26;24(6):1190. doi: 10.3390/molecules24061190. Molecules. 2019. PMID: 30917623 Free PMC article. Review.
Cited by
-
Checkpoint inhibition through small molecule-induced internalization of programmed death-ligand 1.Nat Commun. 2021 Feb 22;12(1):1222. doi: 10.1038/s41467-021-21410-1. Nat Commun. 2021. PMID: 33619272 Free PMC article.
-
Camrelizumab-targeting a novel PD-1 epitope to treat hepatocellular carcinoma.Ann Transl Med. 2020 Dec;8(23):1614. doi: 10.21037/atm-2020-115. Ann Transl Med. 2020. PMID: 33437813 Free PMC article. No abstract available.
-
Comparative Analytical Evaluation of the Proposed Biosimilar FYB206 and its Reference Medicinal Product Keytruda®.Drugs R D. 2024 Sep;24(3):447-464. doi: 10.1007/s40268-024-00485-3. Epub 2024 Sep 4. Drugs R D. 2024. PMID: 39230843 Free PMC article.
-
Passive Membrane Permeability of Sizable Acyclic β-Hairpin Peptides.ACS Med Chem Lett. 2023 Jan 27;14(3):278-284. doi: 10.1021/acsmedchemlett.2c00486. eCollection 2023 Mar 9. ACS Med Chem Lett. 2023. PMID: 36923919 Free PMC article.
-
Variable PD-1 glycosylation modulates the activity of immune checkpoint inhibitors.Life Sci Alliance. 2024 Jan 4;7(3):e202302368. doi: 10.26508/lsa.202302368. Print 2024 Mar. Life Sci Alliance. 2024. PMID: 38176728 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

