Resident memory T cells are a cellular reservoir for HIV in the cervical mucosa
- PMID: 31628331
- PMCID: PMC6802119
- DOI: 10.1038/s41467-019-12732-2
Resident memory T cells are a cellular reservoir for HIV in the cervical mucosa
Abstract
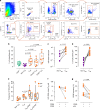
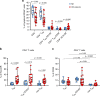
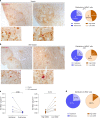
HIV viral reservoirs are established very early during infection. Resident memory T cells (TRM) are present in tissues such as the lower female genital tract, but the contribution of this subset of cells to the pathogenesis and persistence of HIV remains unclear. Here, we show that cervical CD4+TRM display a unique repertoire of clusters of differentiation, with enrichment of several molecules associated with HIV infection susceptibility, longevity and self-renewing capacities. These protein profiles are enriched in a fraction of CD4+TRM expressing CD32. Cervical explant models show that CD4+TRM preferentially support HIV infection and harbor more viral DNA and protein than non-TRM. Importantly, cervical tissue from ART-suppressed HIV+ women contain high levels of viral DNA and RNA, being the TRM fraction the principal contributor. These results recognize the lower female genital tract as an HIV sanctuary and identify CD4+TRM as primary targets of HIV infection and viral persistence. Thus, strategies towards an HIV cure will need to consider TRM phenotypes, which are widely distributed in tissues.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
A Novel Single-Cell FISH-Flow Assay Identifies Effector Memory CD4+ T cells as a Major Niche for HIV-1 Transcription in HIV-Infected Patients.mBio. 2017 Jul 11;8(4):e00876-17. doi: 10.1128/mBio.00876-17. mBio. 2017. PMID: 28698276 Free PMC article.
-
Persistence of an intact HIV reservoir in phenotypically naive T cells.JCI Insight. 2020 Oct 15;5(20):e133157. doi: 10.1172/jci.insight.133157. JCI Insight. 2020. PMID: 33055422 Free PMC article.
-
Effector memory differentiation increases detection of replication-competent HIV-l in resting CD4+ T cells from virally suppressed individuals.PLoS Pathog. 2019 Oct 14;15(10):e1008074. doi: 10.1371/journal.ppat.1008074. eCollection 2019 Oct. PLoS Pathog. 2019. PMID: 31609991 Free PMC article.
-
Blocking Formation of the Stable HIV Reservoir: A New Perspective for HIV-1 Cure.Front Immunol. 2019 Aug 22;10:1966. doi: 10.3389/fimmu.2019.01966. eCollection 2019. Front Immunol. 2019. PMID: 31507594 Free PMC article. Review.
-
The Role of Tissue Resident Memory CD4 T Cells in Herpes Simplex Viral and HIV Infection.Viruses. 2021 Feb 25;13(3):359. doi: 10.3390/v13030359. Viruses. 2021. PMID: 33668777 Free PMC article. Review.
Cited by
-
Differential expression of HIV target cells CCR5 and α4β7 in tissue resident memory CD4 T cells in endocervix during the menstrual cycle of HIV seronegative women.Front Immunol. 2024 Sep 25;15:1456652. doi: 10.3389/fimmu.2024.1456652. eCollection 2024. Front Immunol. 2024. PMID: 39386203 Free PMC article.
-
Sulforaphane prevents the reactivation of HIV-1 by suppressing NFκB signaling.J Virus Erad. 2023 Aug 12;9(3):100341. doi: 10.1016/j.jve.2023.100341. eCollection 2023 Sep. J Virus Erad. 2023. PMID: 37663574 Free PMC article.
-
Tissue-specific differences in HIV DNA levels and mechanisms that govern HIV transcription in blood, gut, genital tract and liver in ART-treated women.J Int AIDS Soc. 2021 Jul;24(7):e25738. doi: 10.1002/jia2.25738. J Int AIDS Soc. 2021. PMID: 34235864 Free PMC article.
-
Intraepithelial CD15 infiltration identifies high-grade anal dysplasia in people with HIV.JCI Insight. 2024 Jun 20;9(15):e175251. doi: 10.1172/jci.insight.175251. JCI Insight. 2024. PMID: 38900571 Free PMC article.
-
Phenotypic analysis of the unstimulated in vivo HIV CD4 T cell reservoir.Elife. 2020 Sep 29;9:e60933. doi: 10.7554/eLife.60933. Elife. 2020. PMID: 32990219 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

