Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs
- PMID: 31591578
- PMCID: PMC6858545
- DOI: 10.1038/s41592-019-0575-8
Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs
Abstract
Cryo-electron microscopy is a popular method for the determination of protein structures; however, identifying a sufficient number of particles for analysis can take months of manual effort. Current computational approaches find many false positives and require ad hoc postprocessing, especially for unusually shaped particles. To address these shortcomings, we develop Topaz, an efficient and accurate particle-picking pipeline using neural networks trained with a general-purpose positive-unlabeled learning method. This framework enables particle detection models to be trained with few sparsely labeled particles and no labeled negatives. Topaz retrieves many more real particles than conventional picking methods while maintaining low false-positive rates, is capable of picking challenging unusually shaped proteins (for example, small, non-globular and asymmetric particles), produces more representative particle sets and does not require post hoc curation. We demonstrate the performance of Topaz on two difficult datasets and three conventional datasets. Topaz is modular, standalone, free and open source ( http://topaz.csail.mit.edu ).
Conflict of interest statement
Competing financial interests
The authors declare no competing financial interests.
Figures
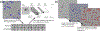


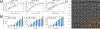
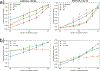
Similar articles
-
AutoCryoPicker: an unsupervised learning approach for fully automated single particle picking in Cryo-EM images.BMC Bioinformatics. 2019 Jun 13;20(1):326. doi: 10.1186/s12859-019-2926-y. BMC Bioinformatics. 2019. PMID: 31195977 Free PMC article.
-
DRPnet: automated particle picking in cryo-electron micrographs using deep regression.BMC Bioinformatics. 2021 Feb 8;22(1):55. doi: 10.1186/s12859-020-03948-x. BMC Bioinformatics. 2021. PMID: 33557750 Free PMC article.
-
CryoTransformer: a transformer model for picking protein particles from cryo-EM micrographs.Bioinformatics. 2024 Mar 4;40(3):btae109. doi: 10.1093/bioinformatics/btae109. Bioinformatics. 2024. PMID: 38407301 Free PMC article.
-
[A review of automatic particle recognition in Cryo-EM images].Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2010 Oct;27(5):1178-82. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2010. PMID: 21089695 Review. Chinese.
-
Review: automatic particle detection in electron microscopy.J Struct Biol. 2001 Feb-Mar;133(2-3):90-101. doi: 10.1006/jsbi.2001.4348. J Struct Biol. 2001. PMID: 11472081 Review.
Cited by
-
Structure of the flotillin complex in a native membrane environment.Proc Natl Acad Sci U S A. 2024 Jul 16;121(29):e2409334121. doi: 10.1073/pnas.2409334121. Epub 2024 Jul 10. Proc Natl Acad Sci U S A. 2024. PMID: 38985763 Free PMC article.
-
Human coronavirus HKU1 recognition of the TMPRSS2 host receptor.bioRxiv [Preprint]. 2024 Jan 9:2024.01.09.574565. doi: 10.1101/2024.01.09.574565. bioRxiv. 2024. Update in: Cell. 2024 Aug 8;187(16):4231-4245.e13. doi: 10.1016/j.cell.2024.06.006 PMID: 38260518 Free PMC article. Updated. Preprint.
-
MiLoPYP: self-supervised molecular pattern mining and particle localization in situ.Nat Methods. 2024 Oct;21(10):1863-1872. doi: 10.1038/s41592-024-02403-6. Epub 2024 Sep 9. Nat Methods. 2024. PMID: 39251798 Free PMC article.
-
Structural basis for accommodation of emerging B.1.351 and B.1.1.7 variants by two potent SARS-CoV-2 neutralizing antibodies.Structure. 2021 Jul 1;29(7):655-663.e4. doi: 10.1016/j.str.2021.05.014. Epub 2021 Jun 9. Structure. 2021. PMID: 34111408 Free PMC article.
-
Stoichiometry and architecture of the human pyruvate dehydrogenase complex.Sci Adv. 2024 Jul 19;10(29):eadn4582. doi: 10.1126/sciadv.adn4582. Epub 2024 Jul 17. Sci Adv. 2024. PMID: 39018392 Free PMC article.
References
-
- Rosenthal PB & Henderson R Optimal Determination of Particle Orientation, Absolute Hand, and Contrast Loss in Single-particle Electron Cryomicroscopy. J. Mol. Bio 333, 721–745 (2003). - PubMed
-
- Tang G et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol 157, 38–46 (2007). - PubMed
Methods-only References
-
- Xu H et al. Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin. Cell 176, 702–715 (2019). - PubMed
-
- Ioffe S & Szegedy C Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. in International Conference on Machine Learning 448–456 (2015).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

