How do DNA-bound proteins leave their binding sites? The role of facilitated dissociation
- PMID: 31586479
- PMCID: PMC6926143
- DOI: 10.1016/j.cbpa.2019.08.007
How do DNA-bound proteins leave their binding sites? The role of facilitated dissociation
Abstract
Dissociation of a protein from DNA is often assumed to be described by an off rate that is independent of other molecules in solution. Recent experiments and computational analyses have challenged this view by showing that unbinding rates (residence times) of DNA-bound proteins can depend on concentrations of nearby molecules that are competing for binding. This 'facilitated dissociation' (FD) process can occur at the single-binding site level via formation of a ternary complex, and can dominate over 'spontaneous dissociation' at low (submicromolar) concentrations. In the crowded intracellular environment FD introduces new regulatory possibilities at the level of individual biomolecule interactions.
Published by Elsevier Ltd.
Figures
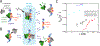
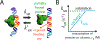

Similar articles
-
Facilitated dissociation of transcription factors from single DNA binding sites.Proc Natl Acad Sci U S A. 2017 Apr 18;114(16):E3251-E3257. doi: 10.1073/pnas.1701884114. Epub 2017 Mar 31. Proc Natl Acad Sci U S A. 2017. PMID: 28364020 Free PMC article.
-
Facilitated Dissociation Kinetics of Dimeric Nucleoid-Associated Proteins Follow a Universal Curve.Biophys J. 2017 Feb 7;112(3):543-551. doi: 10.1016/j.bpj.2016.11.3198. Epub 2016 Dec 21. Biophys J. 2017. PMID: 28012548 Free PMC article.
-
DNA-Segment-Facilitated Dissociation of Fis and NHP6A from DNA Detected via Single-Molecule Mechanical Response.J Mol Biol. 2015 Sep 25;427(19):3123-36. doi: 10.1016/j.jmb.2015.07.015. Epub 2015 Jul 26. J Mol Biol. 2015. PMID: 26220077 Free PMC article.
-
Facilitated Unbinding via Multivalency-Enabled Ternary Complexes: New Paradigm for Protein-DNA Interactions.Acc Chem Res. 2018 Apr 17;51(4):860-868. doi: 10.1021/acs.accounts.7b00541. Epub 2018 Jan 25. Acc Chem Res. 2018. PMID: 29368512 Free PMC article. Review.
-
How do site-specific DNA-binding proteins find their targets?Nucleic Acids Res. 2004 Jun 3;32(10):3040-52. doi: 10.1093/nar/gkh624. Print 2004. Nucleic Acids Res. 2004. PMID: 15178741 Free PMC article. Review.
Cited by
-
The HMGB chromatin protein Nhp6A can bypass obstacles when traveling on DNA.Nucleic Acids Res. 2020 Nov 4;48(19):10820-10831. doi: 10.1093/nar/gkaa799. Nucleic Acids Res. 2020. PMID: 32997109 Free PMC article.
-
Single molecule characterization of the binding kinetics of a transcription factor and its modulation by DNA sequence and methylation.Nucleic Acids Res. 2021 Nov 8;49(19):10975-10987. doi: 10.1093/nar/gkab843. Nucleic Acids Res. 2021. PMID: 34606618 Free PMC article.
-
The Mediator complex as a master regulator of transcription by RNA polymerase II.Nat Rev Mol Cell Biol. 2022 Nov;23(11):732-749. doi: 10.1038/s41580-022-00498-3. Epub 2022 Jun 20. Nat Rev Mol Cell Biol. 2022. PMID: 35725906 Free PMC article. Review.
-
PRC2 direct transfer from G-quadruplex RNA to dsDNA has implications for RNA-binding chromatin modifiers.Proc Natl Acad Sci U S A. 2023 Jun 6;120(23):e2220528120. doi: 10.1073/pnas.2220528120. Epub 2023 May 30. Proc Natl Acad Sci U S A. 2023. PMID: 37252986 Free PMC article.
-
Dynamics of Proteins and Macromolecular Machines in Escherichia coli.EcoSal Plus. 2021 Dec 15;9(2):eESP00112020. doi: 10.1128/ecosalplus.ESP-0011-2020. Epub 2021 Jun 1. EcoSal Plus. 2021. PMID: 34060908 Free PMC article. Review.
References
-
- von Hippel PH Berg OG: Facilitated target location in biological systems. J Biol Chem 1989, 264: 675–678. - PubMed
-
-
Kamar RI, Banigan EJ, Erbaş A, Giuntoli RD, Olvera de la Cruz M, Johnson RC, Marko JF: Facilitated dissociation of transcription factors from single DNA binding sites. Proc Nat Acad Sci USA 2017, 114: E3251–E3257.
•Single-molecule study of FD for Fis and other proteins interacting with a short 27 bp DNA, showing large increase of off rate with increasing free protein concentration, with saturation at large concentration.
-
-
-
Chen TY, Santiago AG, Jung W, Krzemiński Å, Yang F, Martell DJ, Helmann JD, Chen P: Concentration- and chromosome-organization-dependent regulator unbinding from DNA for transcription regulation in living cells. Nat Comm 2015, 6: 7445.
•In vivo observation of FD of a bacterial transcription factor.
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

