Molecular mechanism of TRPV2 channel modulation by cannabidiol
- PMID: 31566564
- PMCID: PMC6794088
- DOI: 10.7554/eLife.48792
Molecular mechanism of TRPV2 channel modulation by cannabidiol
Abstract
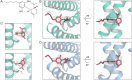
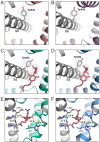
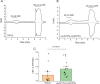
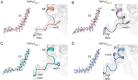
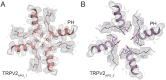
Transient receptor potential vanilloid 2 (TRPV2) plays a critical role in neuronal development, cardiac function, immunity, and cancer. Cannabidiol (CBD), the non-psychotropic therapeutically active ingredient of Cannabis sativa, is an activator of TRPV2 and also modulates other transient receptor potential (TRP) channels. Here, we determined structures of the full-length rat TRPV2 channel in apo and CBD-bound states in nanodiscs by cryo-electron microscopy. We show that CBD interacts with TRPV2 through a hydrophobic pocket located between S5 and S6 helices of adjacent subunits, which differs from known ligand and lipid binding sites in other TRP channels. CBD-bound TRPV2 structures revealed that the S4-S5 linker plays a critical role in channel gating upon CBD binding. Additionally, nanodiscs permitted us to visualize two distinct TRPV2 apo states in a lipid environment. Together these results provide a foundation to further understand TRPV channel gating, their divergent physiological functions, and to accelerate structure-based drug design.
Keywords: S. cerevisiae; TRP channels; cannabidiol; cryo-EM; ion channels; molecular biophysics; structural biology.
© 2019, Pumroy et al.
Conflict of interest statement
RP, AS, TH, SZ, YY, TR, VM No competing interests declared, YL, SH is affiliated with Pfizer Research and Development. The author has no financial interests to declare
Figures











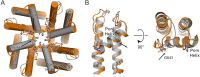
Similar articles
-
Structural insights into TRPV2 activation by small molecules.Nat Commun. 2022 Apr 28;13(1):2334. doi: 10.1038/s41467-022-30083-3. Nat Commun. 2022. PMID: 35484159 Free PMC article.
-
Cannabidiol sensitizes TRPV2 channels to activation by 2-APB.Elife. 2023 May 18;12:e86166. doi: 10.7554/eLife.86166. Elife. 2023. PMID: 37199723 Free PMC article.
-
Structure of the full-length TRPV2 channel by cryo-EM.Nat Commun. 2016 Mar 29;7:11130. doi: 10.1038/ncomms11130. Nat Commun. 2016. PMID: 27021073 Free PMC article.
-
Determining the Crystal Structure of TRPV6.In: Kozak JA, Putney JW Jr, editors. Calcium Entry Channels in Non-Excitable Cells. Boca Raton (FL): CRC Press/Taylor & Francis; 2018. Chapter 14. In: Kozak JA, Putney JW Jr, editors. Calcium Entry Channels in Non-Excitable Cells. Boca Raton (FL): CRC Press/Taylor & Francis; 2018. Chapter 14. PMID: 30299652 Free Books & Documents. Review.
-
Pharmacological effects of cannabidiol by transient receptor potential channels.Life Sci. 2022 Jul 1;300:120582. doi: 10.1016/j.lfs.2022.120582. Epub 2022 Apr 26. Life Sci. 2022. PMID: 35483477 Review.
Cited by
-
Regulation of transient receptor potential channels by traditional Chinese medicines and their active ingredients.Front Pharmacol. 2022 Oct 13;13:1039412. doi: 10.3389/fphar.2022.1039412. eCollection 2022. Front Pharmacol. 2022. PMID: 36313301 Free PMC article. Review.
-
Non-psychotropic phytocannabinoid interactions with voltage-gated sodium channels: An update on cannabidiol and cannabigerol.Front Physiol. 2022 Nov 10;13:1066455. doi: 10.3389/fphys.2022.1066455. eCollection 2022. Front Physiol. 2022. PMID: 36439273 Free PMC article. Review.
-
TRP channels: a journey towards a molecular understanding of pain.Nat Rev Neurosci. 2022 Oct;23(10):596-610. doi: 10.1038/s41583-022-00611-7. Epub 2022 Jul 13. Nat Rev Neurosci. 2022. PMID: 35831443 Review.
-
Molecular pathway and structural mechanism of human oncochannel TRPV6 inhibition by the phytocannabinoid tetrahydrocannabivarin.Nat Commun. 2023 Aug 2;14(1):4630. doi: 10.1038/s41467-023-40362-2. Nat Commun. 2023. PMID: 37532722 Free PMC article.
-
Ligand activation mechanisms of human KCNQ2 channel.Nat Commun. 2023 Oct 19;14(1):6632. doi: 10.1038/s41467-023-42416-x. Nat Commun. 2023. PMID: 37857637 Free PMC article.
References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallographica Section D Biological Crystallography. 2002;58:1948–1954. doi: 10.1107/S0907444902016657. - DOI - PubMed
-
- De Petrocellis L, Vellani V, Schiano-Moriello A, Marini P, Magherini PC, Orlando P, Di Marzo V. Plant-Derived cannabinoids modulate the activity of transient receptor potential channels of ankyrin Type-1 and melastatin Type-8. Journal of Pharmacology and Experimental Therapeutics. 2008;325:1007–1015. doi: 10.1124/jpet.107.134809. - DOI - PubMed
-
- De Petrocellis L, Orlando P, Moriello AS, Aviello G, Stott C, Izzo AA, Di Marzo V. Cannabinoid actions at TRPV channels: effects on TRPV3 and TRPV4 and their potential relevance to gastrointestinal inflammation. Acta Physiologica. 2012;204:255–266. doi: 10.1111/j.1748-1716.2011.02338.x. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

