CYP72A enzymes catalyse 13-hydrolyzation of gibberellins
- PMID: 31527846
- PMCID: PMC7194175
- DOI: 10.1038/s41477-019-0511-z
CYP72A enzymes catalyse 13-hydrolyzation of gibberellins
Erratum in
-
Author Correction: CYP72A enzymes catalyse 13-hydrolyzation of gibberellins.Nat Plants. 2019 Nov;5(11):1194. doi: 10.1038/s41477-019-0531-8. Nat Plants. 2019. PMID: 31582809
Abstract
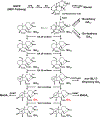
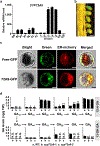
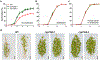
Bioactive gibberellins (GAs or diterpenes) are essential hormones in land plants that control many aspects of plant growth and development. In flowering plants, 13-OH GAs (having low bioactivity-for example, GA1) and 13-H GAs (having high bioactivity-for example, GA4) frequently coexist in the same plant. However, the identity of the native Arabidopsis thaliana 13-hydroxylase GA and its physiological functions remain unknown. Here, we report that cytochrome P450 genes (CYP72A9 and its homologues) encode active GA 13-hydroxylases in Brassicaceae. Plants overexpressing CYP72A9 exhibited semi-dwarfism, which was caused by significant reduction in GA4 levels. Biochemical assays revealed that recombinant CYP72A9 protein catalysed the conversion of 13-H GAs to the corresponding 13-OH GAs. CYP72A9 was expressed predominantly in developing seeds in Arabidopsis. Freshly harvested seeds of cyp72a9 mutants germinated more quickly than the wild type, whereas stratification-treated seeds and seeds from long-term storage did not. The evolutionary origin of GA 13-oxidases from the CYP72A subfamily was also investigated and discussed here.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures



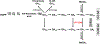
Similar articles
-
CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice.Proc Natl Acad Sci U S A. 2013 Jan 29;110(5):1947-52. doi: 10.1073/pnas.1215788110. Epub 2013 Jan 14. Proc Natl Acad Sci U S A. 2013. PMID: 23319637 Free PMC article.
-
Functional analysis of Arabidopsis CYP714A1 and CYP714A2 reveals that they are distinct gibberellin modification enzymes.Plant Cell Physiol. 2013 Nov;54(11):1837-51. doi: 10.1093/pcp/pct125. Epub 2013 Sep 4. Plant Cell Physiol. 2013. PMID: 24009336
-
High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds.Plant Physiol. 2008 Mar;146(3):1368-85. doi: 10.1104/pp.107.113738. Epub 2007 Dec 27. Plant Physiol. 2008. PMID: 18162586 Free PMC article.
-
Genetic analyses of interactions among gibberellin, abscisic acid, and brassinosteroids in the control of flowering time in Arabidopsis thaliana.PLoS One. 2010 Nov 17;5(11):e14012. doi: 10.1371/journal.pone.0014012. PLoS One. 2010. PMID: 21103336 Free PMC article.
-
Regulation of gibberellin biosynthesis by light.Curr Opin Plant Biol. 1999 Oct;2(5):398-403. doi: 10.1016/s1369-5266(99)00012-6. Curr Opin Plant Biol. 1999. PMID: 10508758 Review.
Cited by
-
The fine mapping of dwarf gene Rht5 in bread wheat and its effects on plant height and main agronomic traits.Planta. 2022 May 4;255(6):114. doi: 10.1007/s00425-022-03888-1. Planta. 2022. PMID: 35507093
-
Occurrence and biosynthesis of plant sesterterpenes (C25), a new addition to terpene diversity.Plant Commun. 2021 Mar 30;2(5):100184. doi: 10.1016/j.xplc.2021.100184. eCollection 2021 Sep 13. Plant Commun. 2021. PMID: 34746758 Free PMC article. Review.
-
Ectopic Expression of the Transcriptional Regulator silky3 Causes Pleiotropic Meristem and Sex Determination Defects in Maize Inflorescences.Plant Cell. 2020 Dec;32(12):3750-3773. doi: 10.1105/tpc.20.00043. Epub 2020 Sep 28. Plant Cell. 2020. PMID: 32989171 Free PMC article.
-
Biosynthetic Pathways of Hormones in Plants.Metabolites. 2023 Jul 25;13(8):884. doi: 10.3390/metabo13080884. Metabolites. 2023. PMID: 37623827 Free PMC article. Review.
-
Highlights in gibberellin research: A tale of the dwarf and the slender.Plant Physiol. 2024 Apr 30;195(1):111-134. doi: 10.1093/plphys/kiae044. Plant Physiol. 2024. PMID: 38290048 Free PMC article. Review.
References
-
- Yang YY et al. Effects of gibberellins on seed germination of phytochrome deficient mutants of Arabidopsis thaliana. Plant Cell Physiol. 36, 1205–1211 (1995). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

