Coronavirus genomic RNA packaging
- PMID: 31505321
- PMCID: PMC7112113
- DOI: 10.1016/j.virol.2019.08.031
Coronavirus genomic RNA packaging
Abstract
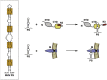
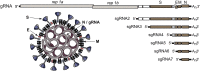
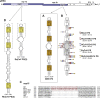
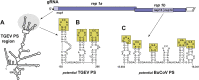
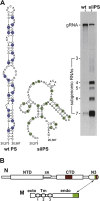
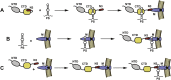
RNA viruses carry out selective packaging of their genomes in a variety of ways, many involving a genomic packaging signal. The first coronavirus packaging signal was discovered nearly thirty years ago, but how it functions remains incompletely understood. This review addresses the current state of knowledge of coronavirus genome packaging, which has mainly been studied in two prototype species, mouse hepatitis virus and transmissible gastroenteritis virus. Despite the progress that has been made in the mapping and characterization of some packaging signals, there is conflicting evidence as to whether the viral nucleocapsid protein or the membrane protein plays the primary role in packaging signal recognition. The different models for the mechanism of genomic RNA packaging that have been prompted by these competing views are described. Also discussed is the recent exciting discovery that selective coronavirus genome packaging is critical for in vivo evasion of the host innate immune response.
Keywords: Coronavirus; Innate immunity; Membrane protein; Mouse hepatitis virus; Nucleocapsid protein; Packaging signal; RNA virus; Viral genome packaging.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Recognition of the murine coronavirus genomic RNA packaging signal depends on the second RNA-binding domain of the nucleocapsid protein.J Virol. 2014 Apr;88(8):4451-65. doi: 10.1128/JVI.03866-13. Epub 2014 Feb 5. J Virol. 2014. PMID: 24501403 Free PMC article.
-
Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal.J Virol. 2003 Mar;77(5):2922-7. doi: 10.1128/jvi.77.5.2922-2927.2003. J Virol. 2003. PMID: 12584316 Free PMC article.
-
Selective Packaging in Murine Coronavirus Promotes Virulence by Limiting Type I Interferon Responses.mBio. 2018 May 1;9(3):e00272-18. doi: 10.1128/mBio.00272-18. mBio. 2018. PMID: 29717007 Free PMC article.
-
RNA Encapsidation and Packaging in the Phleboviruses.Viruses. 2016 Jul 15;8(7):194. doi: 10.3390/v8070194. Viruses. 2016. PMID: 27428993 Free PMC article. Review.
-
Nucleocapsid proteins: roles beyond viral RNA packaging.Wiley Interdiscip Rev RNA. 2016 Mar-Apr;7(2):213-26. doi: 10.1002/wrna.1326. Epub 2016 Jan 8. Wiley Interdiscip Rev RNA. 2016. PMID: 26749541 Free PMC article. Review.
Cited by
-
The molecular virology of coronaviruses.J Biol Chem. 2020 Sep 11;295(37):12910-12934. doi: 10.1074/jbc.REV120.013930. Epub 2020 Jul 13. J Biol Chem. 2020. PMID: 32661197 Free PMC article. Review.
-
Knowledge and attitude towards Covid-19 vaccine in Ethiopia: Systematic review and meta-analysis.Hum Vaccin Immunother. 2023 Dec 31;19(1):2179224. doi: 10.1080/21645515.2023.2179224. Epub 2023 Mar 7. Hum Vaccin Immunother. 2023. PMID: 36882983 Free PMC article.
-
Performance characteristics of the boson rapid SARS-cov-2 antigen test card vs RT-PCR: Cross-reactivity and emerging variants.Heliyon. 2023 Feb;9(2):e13642. doi: 10.1016/j.heliyon.2023.e13642. Epub 2023 Feb 10. Heliyon. 2023. PMID: 36789386 Free PMC article.
-
Current Understanding of Molecular Phase Separation in Chromosomes.Int J Mol Sci. 2021 Oct 4;22(19):10736. doi: 10.3390/ijms221910736. Int J Mol Sci. 2021. PMID: 34639077 Free PMC article. Review.
-
Targeting SARS-CoV-2 Macrodomain-1 to Restore the Innate Immune Response Using In Silico Screening of Medicinal Compounds and Free Energy Calculation Approaches.Viruses. 2023 Sep 12;15(9):1907. doi: 10.3390/v15091907. Viruses. 2023. PMID: 37766313 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

