X Chromosome Domain Architecture Regulates Caenorhabditis elegans Lifespan but Not Dosage Compensation
- PMID: 31495695
- PMCID: PMC6810858
- DOI: 10.1016/j.devcel.2019.08.004
X Chromosome Domain Architecture Regulates Caenorhabditis elegans Lifespan but Not Dosage Compensation
Abstract
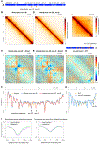
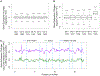
Mechanisms establishing higher-order chromosome structures and their roles in gene regulation are elusive. We analyzed chromosome architecture during nematode X chromosome dosage compensation, which represses transcription via a dosage-compensation condensin complex (DCC) that binds hermaphrodite Xs and establishes megabase-sized topologically associating domains (TADs). We show that DCC binding at high-occupancy sites (rex sites) defines eight TAD boundaries. Single rex deletions disrupted boundaries, and single insertions created new boundaries, demonstrating that a rex site is necessary and sufficient to define DCC-dependent boundary locations. Deleting eight rex sites (8rexΔ) recapitulated TAD structure of DCC mutants, permitting analysis when chromosome-wide domain architecture was disrupted but most DCC binding remained. 8rexΔ animals exhibited no changes in X expression and lacked dosage-compensation mutant phenotypes. Hence, TAD boundaries are neither the cause nor the consequence of DCC-mediated gene repression. Abrogating TAD structure did, however, reduce thermotolerance, accelerate aging, and shorten lifespan, implicating chromosome architecture in stress responses and aging.
Keywords: X chromosome dosage compensation; aging; condensin; gene expression; higher-order chromosome structure; lifespan; proteotoxic stress; topologically associating domains.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures




Comment in
-
TADs for Life: Chromatin Domain Organization Regulates Lifespan in C. elegans.Dev Cell. 2019 Oct 21;51(2):131-132. doi: 10.1016/j.devcel.2019.09.021. Dev Cell. 2019. PMID: 31639364
Similar articles
-
Condensin-driven remodelling of X chromosome topology during dosage compensation.Nature. 2015 Jul 9;523(7559):240-4. doi: 10.1038/nature14450. Epub 2015 Jun 1. Nature. 2015. PMID: 26030525 Free PMC article.
-
An MLL/COMPASS subunit functions in the C. elegans dosage compensation complex to target X chromosomes for transcriptional regulation of gene expression.Genes Dev. 2011 Mar 1;25(5):499-515. doi: 10.1101/gad.2016011. Genes Dev. 2011. PMID: 21363964 Free PMC article.
-
SUMOylation is essential for sex-specific assembly and function of the Caenorhabditis elegans dosage compensation complex on X chromosomes.Proc Natl Acad Sci U S A. 2013 Oct 1;110(40):E3810-9. doi: 10.1073/pnas.1315793110. Epub 2013 Sep 16. Proc Natl Acad Sci U S A. 2013. PMID: 24043781 Free PMC article.
-
Caenorhabditis elegans Dosage Compensation: Insights into Condensin-Mediated Gene Regulation.Trends Genet. 2018 Jan;34(1):41-53. doi: 10.1016/j.tig.2017.09.010. Epub 2017 Oct 13. Trends Genet. 2018. PMID: 29037439 Free PMC article. Review.
-
Targeting X chromosomes for repression.Curr Opin Genet Dev. 2010 Apr;20(2):179-89. doi: 10.1016/j.gde.2010.03.008. Epub 2010 Apr 8. Curr Opin Genet Dev. 2010. PMID: 20381335 Free PMC article. Review.
Cited by
-
Nuclear genome organization in fungi: from gene folding to Rabl chromosomes.FEMS Microbiol Rev. 2023 May 19;47(3):fuad021. doi: 10.1093/femsre/fuad021. FEMS Microbiol Rev. 2023. PMID: 37197899 Free PMC article.
-
Histone post-translational modifications - cause and consequence of genome function.Nat Rev Genet. 2022 Sep;23(9):563-580. doi: 10.1038/s41576-022-00468-7. Epub 2022 Mar 25. Nat Rev Genet. 2022. PMID: 35338361 Review.
-
Cohesin organizes 3D DNA contacts surrounding active enhancers in C. elegans.bioRxiv [Preprint]. 2024 Sep 25:2023.09.18.558239. doi: 10.1101/2023.09.18.558239. bioRxiv. 2024. PMID: 37786717 Free PMC article. Preprint.
-
Regulation of organelle size and organization during development.Semin Cell Dev Biol. 2023 Jan 15;133:53-64. doi: 10.1016/j.semcdb.2022.02.002. Epub 2022 Feb 8. Semin Cell Dev Biol. 2023. PMID: 35148938 Free PMC article. Review.
-
Transcription-independent TFIIIC-bound sites cluster near heterochromatin boundaries within lamina-associated domains in C. elegans.Epigenetics Chromatin. 2020 Jan 9;13(1):1. doi: 10.1186/s13072-019-0325-2. Epigenetics Chromatin. 2020. PMID: 31918747 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

