Prognostic value and immune infiltration of novel signatures in clear cell renal cell carcinoma microenvironment
- PMID: 31493764
- PMCID: PMC6756904
- DOI: 10.18632/aging.102233
Prognostic value and immune infiltration of novel signatures in clear cell renal cell carcinoma microenvironment
Abstract
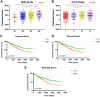
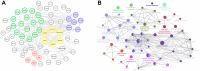
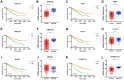
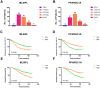
Growing evidence has highlighted the immune response as an important feature of carcinogenesis and therapeutic efficacy in clear cell renal cell carcinoma (ccRCC). This study categorized ccRCC cases into high and low score groups based on their immune/stromal scores generated by the ESTIMATE algorithm, and identified an association between these scores and prognosis. Differentially expressed tumor environment (TME)-related genes extracted from common upregulated components in immune and stromal scores were described using functional annotations and protein-protein interaction (PPI) networks. Most PPIs were selected for further prognostic investigation. Many additional previously neglected signatures, including AGPAT9, AQP7, HMGCS2, KLF15, MLXIPL, PPARGC1A, exhibited significant prognostic potential. In addition, multivariate Cox analysis indicated that MIXIPL and PPARGC1A were the most significant prognostic signatures, and were closely related to immune infiltration in TCGA cohort. External prognostic validation of MIXIPL and PPARGC1A was undertaken in 380 ccRCC cases from a real-world cohort. These findings indicate the relevance of monitoring and manipulation of the microenvironment for ccRCC prognosis and precision immunotherapy.
Keywords: ESTIMATE algorithm; clear cell renal cell carcinoma; immune signature; prognosis; tumor microenvironment.
Conflict of interest statement
Figures

Similar articles
-
Comprehensive insights on pivotal prognostic signature involved in clear cell renal cell carcinoma microenvironment using the ESTIMATE algorithm.Cancer Med. 2020 Jun;9(12):4310-4323. doi: 10.1002/cam4.2983. Epub 2020 Apr 20. Cancer Med. 2020. PMID: 32311223 Free PMC article.
-
Data Mining of Prognostic Microenvironment-Related Genes in Clear Cell Renal Cell Carcinoma: A Study with TCGA Database.Dis Markers. 2019 Oct 30;2019:8901649. doi: 10.1155/2019/8901649. eCollection 2019. Dis Markers. 2019. PMID: 31781309 Free PMC article.
-
Construction of an immune-related prognostic model by exploring the tumor microenvironment of clear cell renal cell carcinoma.Anal Biochem. 2022 Apr 15;643:114567. doi: 10.1016/j.ab.2022.114567. Epub 2022 Feb 3. Anal Biochem. 2022. PMID: 35122734
-
Diagnostic and prognostic tissuemarkers in clear cell and papillary renal cell carcinoma.Cancer Biomark. 2010;7(6):261-8. doi: 10.3233/CBM-2010-0195. Cancer Biomark. 2010. PMID: 21694464 Review.
-
The tumour microenvironment and metabolism in renal cell carcinoma targeted or immune therapy.J Cell Physiol. 2021 Mar;236(3):1616-1627. doi: 10.1002/jcp.29969. Epub 2020 Aug 11. J Cell Physiol. 2021. PMID: 32783202 Review.
Cited by
-
Potential Prognostic Value and Mechanism of Stromal-Immune Signature in Tumor Microenvironment for Stomach Adenocarcinoma.Biomed Res Int. 2020 Jun 26;2020:4673153. doi: 10.1155/2020/4673153. eCollection 2020. Biomed Res Int. 2020. PMID: 32685487 Free PMC article.
-
Role of copper ionophore-induced death in immune microenvironment and clinical prognosis of ccRCC: An integrated analysis.Front Genet. 2022 Oct 3;13:994999. doi: 10.3389/fgene.2022.994999. eCollection 2022. Front Genet. 2022. PMID: 36263424 Free PMC article.
-
The dysfunctional immune response in renal cell carcinoma correlates with changes in the metabolic landscape of ccRCC during disease progression.Cancer Immunol Immunother. 2023 Dec;72(12):4221-4234. doi: 10.1007/s00262-023-03558-5. Epub 2023 Nov 8. Cancer Immunol Immunother. 2023. PMID: 37940720 Free PMC article.
-
Clinical neutrophil-associated genes as reliable predictors of hepatocellular carcinoma.Front Genet. 2022 Oct 6;13:989779. doi: 10.3389/fgene.2022.989779. eCollection 2022. Front Genet. 2022. PMID: 36276937 Free PMC article.
-
Bioinformatic Identification of Neuroblastoma Microenvironment-Associated Biomarkers with Prognostic Value.J Oncol. 2020 Sep 10;2020:5943014. doi: 10.1155/2020/5943014. eCollection 2020. J Oncol. 2020. PMID: 32963529 Free PMC article.
References
-
- Linehan WM, Schmidt LS, Crooks DR, Wei D, Srinivasan R, Lang M, Ricketts CJ. The Metabolic Basis of Kidney Cancer. Cancer Discov. 2019; 9:1006–21. 10.1158/2159-8290.CD-18-1354 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

