Strigolactone synthesis is ancestral in land plants, but canonical strigolactone signalling is a flowering plant innovation
- PMID: 31488154
- PMCID: PMC6728956
- DOI: 10.1186/s12915-019-0689-6
Strigolactone synthesis is ancestral in land plants, but canonical strigolactone signalling is a flowering plant innovation
Abstract
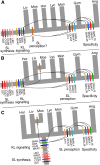
Background: Strigolactones (SLs) are an important class of carotenoid-derived signalling molecule in plants, which function both as exogenous signals in the rhizosphere and as endogenous plant hormones. In flowering plants, SLs are synthesized by a core pathway of four enzymes and are perceived by the DWARF14 (D14) receptor, leading to degradation of SMAX1-LIKE7 (SMXL7) target proteins in a manner dependent on the SCFMAX2 ubiquitin ligase. The evolutionary history of SLs is poorly understood, and it is not clear whether SL synthesis and signalling are present in all land plant lineages, nor when these traits evolved.
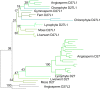
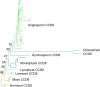
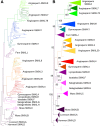
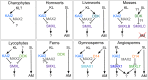
Results: We have utilized recently-generated genomic and transcriptomic sequences from across the land plant clade to resolve the origin of each known component of SL synthesis and signalling. We show that all enzymes in the core SL synthesis pathway originated at or before the base of land plants, consistent with the previously observed distribution of SLs themselves in land plant lineages. We also show that the late-acting enzyme LATERAL BRANCHING OXIDOREDUCTASE (LBO) may be considerably more ancient than previously thought. We perform a detailed phylogenetic analysis of SMXL proteins and show that specific SL target proteins only arose in flowering plants. We also assess diversity and protein structure in the SMXL family, identifying several previously unknown clades.
Conclusions: Overall, our results suggest that SL synthesis is much more ancient than canonical SL signalling, consistent with the idea that SLs first evolved as rhizosphere signals and were only recruited much later as hormonal signals.
Keywords: Phylogenetics; Strigolactone signalling; Strigolactone synthesis; Strigolactones.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Evolution of strigolactone receptors by gradual neo-functionalization of KAI2 paralogues.BMC Biol. 2017 Jun 29;15(1):52. doi: 10.1186/s12915-017-0397-z. BMC Biol. 2017. PMID: 28662667 Free PMC article.
-
Molecular evolution and diversification of the SMXL gene family.J Exp Bot. 2018 Apr 23;69(9):2367-2378. doi: 10.1093/jxb/ery097. J Exp Bot. 2018. PMID: 29538714
-
Fellowship of the rings: a saga of strigolactones and other small signals.New Phytol. 2020 Jan;225(2):621-636. doi: 10.1111/nph.16135. Epub 2019 Sep 17. New Phytol. 2020. PMID: 31442309 Review.
-
Structural modelling and transcriptional responses highlight a clade of PpKAI2-LIKE genes as candidate receptors for strigolactones in Physcomitrella patens.Planta. 2016 Jun;243(6):1441-53. doi: 10.1007/s00425-016-2481-y. Epub 2016 Mar 15. Planta. 2016. PMID: 26979323
-
Novel insights into strigolactone distribution and signalling.Curr Opin Plant Biol. 2013 Oct;16(5):583-9. doi: 10.1016/j.pbi.2013.06.007. Epub 2013 Jul 3. Curr Opin Plant Biol. 2013. PMID: 23830996 Review.
Cited by
-
Genome-Wide Identification of SMXL Gene Family in Soybean and Expression Analysis of GmSMXLs under Shade Stress.Plants (Basel). 2022 Sep 15;11(18):2410. doi: 10.3390/plants11182410. Plants (Basel). 2022. PMID: 36145811 Free PMC article.
-
Karrikin Signaling Acts Parallel to and Additively with Strigolactone Signaling to Regulate Rice Mesocotyl Elongation in Darkness.Plant Cell. 2020 Sep;32(9):2780-2805. doi: 10.1105/tpc.20.00123. Epub 2020 Jul 14. Plant Cell. 2020. PMID: 32665307 Free PMC article.
-
New Paradigms in Brassinosteroids, Strigolactones, Sphingolipids, and Nitric Oxide Interaction in the Control of Lateral and Adventitious Root Formation.Plants (Basel). 2023 Jan 16;12(2):413. doi: 10.3390/plants12020413. Plants (Basel). 2023. PMID: 36679126 Free PMC article. Review.
-
Ancestral sequence reconstruction of the CYP711 family reveals functional divergence in strigolactone biosynthetic enzymes associated with gene duplication events in monocot grasses.New Phytol. 2022 Sep;235(5):1900-1912. doi: 10.1111/nph.18285. Epub 2022 Jun 25. New Phytol. 2022. PMID: 35644901 Free PMC article.
-
Asymmetric expansions of FT and TFL1 lineages characterize differential evolution of the EuPEBP family in the major angiosperm lineages.BMC Biol. 2021 Aug 31;19(1):181. doi: 10.1186/s12915-021-01128-8. BMC Biol. 2021. PMID: 34465318 Free PMC article.
References
-
- López-Ráez JA, Charnikhova T, Gómez-Roldán V, Matusova R, Kohlen W, De Vos R, Verstappen F, Puech-Pages V, Bécard G, Mulder P, Bouwmeester H. Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol. 2008;178:863–874. doi: 10.1111/j.1469-8137.2008.02406.x. - DOI - PubMed
-
- Kohlen W, Charnikhova T, Liu Q, Domagalska MA BR, Beguerie S, Verstappen F, Leyser O, Bouwmeester H, Ruyter-Spira C. Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol. 2011;155:974–987. doi: 10.1104/pp.110.164640. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

