Defining the Identity and Dynamics of Adult Gastric Isthmus Stem Cells
- PMID: 31422913
- PMCID: PMC6739486
- DOI: 10.1016/j.stem.2019.07.008
Defining the Identity and Dynamics of Adult Gastric Isthmus Stem Cells
Abstract
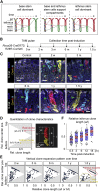
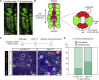
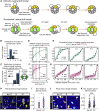
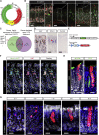
The gastric corpus epithelium is the thickest part of the gastrointestinal tract and is rapidly turned over. Several markers have been proposed for gastric corpus stem cells in both isthmus and base regions. However, the identity of isthmus stem cells (IsthSCs) and the interaction between distinct stem cell populations is still under debate. Here, based on unbiased genetic labeling and biophysical modeling, we show that corpus glands are compartmentalized into two independent zones, with slow-cycling stem cells maintaining the base and actively cycling stem cells maintaining the pit-isthmus-neck region through a process of "punctuated" neutral drift dynamics. Independent lineage tracing based on Stmn1 and Ki67 expression confirmed that rapidly cycling IsthSCs maintain the pit-isthmus-neck region. Finally, single-cell RNA sequencing (RNA-seq) analysis is used to define the molecular identity and lineage relationship of a single, cycling, IsthSC population. These observations define the identity and functional behavior of IsthSCs.
Keywords: Lgr5; Troy; biophysical modeling; deep tissue imaging; gastric corpus isthmus stem cell; intestine; punctuated neutral drift; single-cell RNA-seq; two stem cell compartments; unbiased genetic labeling.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Identification of Stem Cells in the Epithelium of the Stomach Corpus and Antrum of Mice.Gastroenterology. 2017 Jan;152(1):218-231.e14. doi: 10.1053/j.gastro.2016.09.018. Epub 2016 Sep 23. Gastroenterology. 2017. PMID: 27670082
-
Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium.Cell. 2013 Oct 10;155(2):357-68. doi: 10.1016/j.cell.2013.09.008. Cell. 2013. PMID: 24120136 Free PMC article.
-
Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro.Cell Stem Cell. 2010 Jan 8;6(1):25-36. doi: 10.1016/j.stem.2009.11.013. Cell Stem Cell. 2010. PMID: 20085740
-
Adult gastric stem cells and their niches.Wiley Interdiscip Rev Dev Biol. 2017 Mar;6(2). doi: 10.1002/wdev.261. Epub 2017 Jan 3. Wiley Interdiscip Rev Dev Biol. 2017. PMID: 28044412 Review.
-
Clonal Evolution of Stem Cells in the Gastrointestinal Tract.Adv Exp Med Biol. 2016;908:11-25. doi: 10.1007/978-3-319-41388-4_2. Adv Exp Med Biol. 2016. PMID: 27573765 Review.
Cited by
-
Regulation of Parietal Cell Homeostasis by Bone Morphogenetic Protein Signaling.Gastro Hep Adv. 2022 Oct 14;2(2):221-231. doi: 10.1016/j.gastha.2022.10.002. eCollection 2023. Gastro Hep Adv. 2022. PMID: 39132621 Free PMC article.
-
Stem Cell Biomarkers and Tumorigenesis in Gastric Cancer.J Pers Med. 2022 Jun 3;12(6):929. doi: 10.3390/jpm12060929. J Pers Med. 2022. PMID: 35743714 Free PMC article. Review.
-
GPR30-Expressing Gastric Chief Cells Do Not Dedifferentiate But Are Eliminated via PDK-Dependent Cell Competition During Development of Metaplasia.Gastroenterology. 2020 May;158(6):1650-1666.e15. doi: 10.1053/j.gastro.2020.01.046. Epub 2020 Feb 4. Gastroenterology. 2020. PMID: 32032583 Free PMC article.
-
Gastric epithelial histology and precancerous conditions.World J Gastrointest Oncol. 2022 Feb 15;14(2):396-412. doi: 10.4251/wjgo.v14.i2.396. World J Gastrointest Oncol. 2022. PMID: 35317321 Free PMC article. Review.
-
Paligenosis: Cellular Remodeling During Tissue Repair.Annu Rev Physiol. 2022 Feb 10;84:461-483. doi: 10.1146/annurev-physiol-061121-035954. Epub 2021 Oct 27. Annu Rev Physiol. 2022. PMID: 34705482 Free PMC article. Review.
References
-
- Bartfeld S., Koo B.K. Adult gastric stem cells and their niches. Wiley Interdisc. Rev. Dev. Biol. 2017;6:e261. - PubMed
-
- Basak O., Krieger T.G., Muraro M.J., Wiebrands K., Stange D.E., Frias-Aldeguer J., Rivron N.C., van de Wetering M., van Es J.H., van Oudenaarden A. Troy+ brain stem cells cycle through quiescence and regulate their number by sensing niche occupancy. Proc. Natl. Acad. Sci. USA. 2018;115:E610–E619. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

